Differential stability of the bovine prion protein upon urea unfolding
- PMID: 19693935
- PMCID: PMC2786980
- DOI: 10.1002/pro.231
Differential stability of the bovine prion protein upon urea unfolding
Abstract
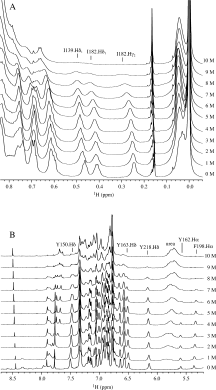
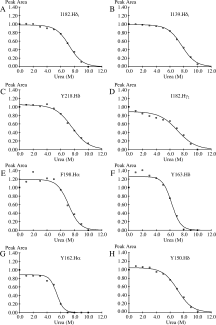
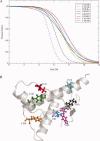
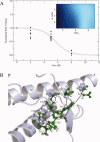
Prion diseases, or transmissible spongiform encephalopathies, are a group of infectious neurological diseases associated with the structural conversion of an endogenous protein (PrP) in the central nervous system. There are two major forms of this protein: the native and noninfectious cellular form, PrP(C); and the misfolded, infectious, and proteinase K-resistant form, PrP(Sc). The C-terminal domain of PrP(C) is mainly alpha-helical in structure, whereas PrP(Sc) in known to aggregate into an assembly of beta-sheets, forming amyloid fibrils. To identify the regions of PrP(C) potentially involved in the initial steps of the conversion to the infectious conformation, we have used high-resolution NMR spectroscopy to characterize the stability and structure of bovine recombinant PrP(C) (residues 121 to 230) during unfolding with the denaturant urea. Analysis of the 800 MHz (1)H NMR spectra reveals region-specific information about the structural changes occurring upon unfolding. Our data suggest that the dissociation of the native beta-sheet of PrP(C) is a primary step in the urea-induced unfolding process, while strong hydrophobic interactions between helices alpha1 and alpha3, and between alpha2 and alpha3, stabilize these regions even at very high concentrations of urea.
Figures



References
-
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. - PubMed
-
- Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. - PubMed
-
- Weissmann C. The state of the prion. Nat Rev Microbiol. 2004;2:861–871. - PubMed
-
- Caughey B, Baron GS. Prions and their partners in crime. Nature. 2006;443:803–810. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

