Cassava mosaic geminiviruses: actual knowledge and perspectives
- PMID: 19694957
- PMCID: PMC6640248
- DOI: 10.1111/j.1364-3703.2009.00559.x
Cassava mosaic geminiviruses: actual knowledge and perspectives
Abstract
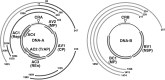
SUMMARY Cassava mosaic disease (CMD) caused by cassava mosaic geminiviruses (CMGs) is one of the most devastating crop diseases and a major constraint for cassava cultivation. CMD has been reported only from the African continent and Indian subcontinent despite the large-scale cultivation of cassava in Latin America and several South-East Asian countries. Seven CMG species have been reported from Africa and two from the Indian subcontinent and, in addition, several strains have been recognized. Recombination and pseudo-recombination between CMGs give rise not only to different strains, but also to members of novel virus species with increased virulence and a new source of biodiversity, causing severe disease epidemics. CMGs are known to trigger gene silencing in plants and, in order to counteract this natural host defence, geminiviruses have evolved suppressor proteins. Temperature and other environmental factors can affect silencing and suppression, and thus modulate the symptoms. In the case of mixed infections of two or more CMGs, there is a possibility for a synergistic interaction as a result of the presence of differential and combinatorial suppressor proteins. In this article, we provide the status of recent research findings with regard to the CMD complex, present the molecular biology knowledge of CMGs with reference to other geminiviruses, and highlight the mechanisms by which CMGs have exploited nature to their advantage.
Figures





References
-
- Abraham, A. (1956) Tapioca Cultivation in India. Pusa, New Delhi: Indian Council of Agricultural Research.
-
- Akano, O. , Dixon, O. , Mba, C. , Barrera, E. and Fregene, M. (2002) Genetic mapping of a dominant gene conferring resistance to cassava mosaic disease. Theor. Appl. Genet. 105, 521–525. - PubMed
-
- Akbergenov, R. , Si‐Ammour, A. , Blevins, T. , Amin, I. , Kutter, C. , Vanderschuren, H. , Zhang, P. , Gruissem, W. , Meins, F., Jr , Hohn, T. and Pooggin, M.M. (2006) Molecular characterization of geminivirus‐derived small RNAs in different plant species. Nucleic Acids Res. 34, 462–471. - PMC - PubMed
-
- Alagianagalingam, M.N. and Ramakrishnan, K. (1966) Cassava mosaic in India. South Ind. Hort. 14, 71–72.
-
- Alicai, T. , Omongo, C.A. , Maruthi, M.N. , Hillocks, R.J. , Baguma, Y. , Kawuki, R. , Bua, A. , Otim‐Nape, G.W. and Colvin, J. (2007) Re‐emergence of cassava brown streak disease in Uganda. Plant Dis. 91, 24–29. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

