Differential contribution of the m7G-cap to the 5' end-dependent translation initiation of mammalian mRNAs
- PMID: 19696074
- PMCID: PMC2764426
- DOI: 10.1093/nar/gkp665
Differential contribution of the m7G-cap to the 5' end-dependent translation initiation of mammalian mRNAs
Abstract
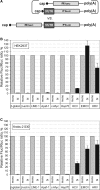
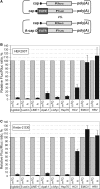
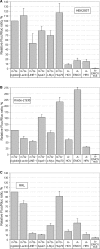
Many mammalian mRNAs possess long 5' UTRs with numerous stem-loop structures. For some of them, the presence of Internal Ribosome Entry Sites (IRESes) was suggested to explain their significant activity, especially when cap-dependent translation is compromised. To test this hypothesis, we have compared the translation initiation efficiencies of some cellular 5' UTRs reported to have IRES-activity with those lacking IRES-elements in RNA-transfected cells and cell-free systems. Unlike viral IRESes, the tested 5' UTRs with so-called 'cellular IRESes' demonstrate only background activities when placed in the intercistronic position of dicistronic RNAs. In contrast, they are very active in the monocistronic context and the cap is indispensable for their activities. Surprisingly, in cultured cells or cytoplasmic extracts both the level of stimulation with the cap and the overall translation activity do not correlate with the cumulative energy of the secondary structure of the tested 5' UTRs. The cap positive effect is still observed under profound inhibition of translation with eIF4E-BP1 but its magnitude varies for individual 5' UTRs irrespective of the cumulative energy of their secondary structures. Thus, it is not mandatory to invoke the IRES hypothesis, at least for some mRNAs, to explain their preferential translation when eIF4E is partially inactivated.
Figures



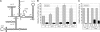
References
-
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

