CD4-CD8 lineage differentiation: Thpok-ing into the nucleus
- PMID: 19696430
- PMCID: PMC3387994
- DOI: 10.4049/jimmunol.0901041
CD4-CD8 lineage differentiation: Thpok-ing into the nucleus
Abstract
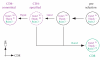
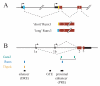
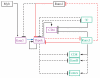
The mature alphabeta T cell population is divided into two main lineages that are defined by the mutually exclusive expression of CD4 and CD8 surface molecules (coreceptors) and that differ in their MHC restriction and function. CD4 T cells are typically MHC-II restricted and helper (or regulatory), whereas CD8 T cells are typically cytotoxic. Several transcription factors are known to control the emergence of CD4 and CD8 lineages, including the zinc finger proteins Thpok and Gata3, which are required for CD4 lineage differentiation, and the Runx factors Runx1 and Runx3, which contribute to CD8 lineage differentiation. This review summarizes recent advances on the function of these transcription factors in lineage differentiation. We also discuss how the "circuitry" connecting these factors could operate to match the expression of the lineage-committing factors Thpok and Runx3, and therefore lineage differentiation, to MHC specificity.
Figures
References
-
- Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. - PubMed
-
- Teh HS, Kisielow P, Scott B, Kishi H, Uematsu Y, Bluthmann H, von Boehmer H. Thymic major histocompatibility complex antigens and the alpha beta T-cell receptor determine the CD4/CD8 phenotype of T cells. Nature. 1988;335:229–233. - PubMed
-
- Sha WC, Nelson CA, Newberry RD, Kranz DM, Russell JH, Loh DY. Selective expression of an antigen receptor on CD8-bearing T lymphocytes in transgenic mice. Nature. 1988;335:271–274. - PubMed
-
- Kaye J, Hsu ML, Sauron ME, Jameson SC, Gascoigne NR, Hedrick SM. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 1989;341:746–749. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

