Allosteric beta-propeller signalling in TolB and its manipulation by translocating colicins
- PMID: 19696740
- PMCID: PMC2750012
- DOI: 10.1038/emboj.2009.224
Allosteric beta-propeller signalling in TolB and its manipulation by translocating colicins
Erratum in
- EMBO J. 2009 Sep 16;28(18):2858
Abstract
The Tol system is a five-protein assembly parasitized by colicins and bacteriophages that helps stabilize the Gram-negative outer membrane (OM). We show that allosteric signalling through the six-bladed beta-propeller protein TolB is central to Tol function in Escherichia coli and that this is subverted by colicins such as ColE9 to initiate their OM translocation. Protein-protein interactions with the TolB beta-propeller govern two conformational states that are adopted by the distal N-terminal 12 residues of TolB that bind TolA in the inner membrane. ColE9 promotes disorder of this 'TolA box' and recruitment of TolA. In contrast to ColE9, binding of the OM lipoprotein Pal to the same site induces conformational changes that sequester the TolA box to the TolB surface in which it exhibits little or no TolA binding. Our data suggest that Pal is an OFF switch for the Tol assembly, whereas colicins promote an ON state even though mimicking Pal. Comparison of the TolB mechanism to that of vertebrate guanine nucleotide exchange factor RCC1 suggests that allosteric signalling may be more prevalent in beta-propeller proteins than currently realized.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

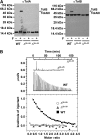
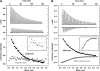
 ). The cell concentration of the starting complex was 60 μM into which was injected TolAIII. No TolAIII binding could be detected when Pal was bound to TolB. (B) Raw ITC data, top panel and integrated heats of binding for the TolB–TolAIII complex (▪) and the corresponding data when TolB was complexed with the ColE9 T-domain (
). The cell concentration of the starting complex was 60 μM into which was injected TolAIII. No TolAIII binding could be detected when Pal was bound to TolB. (B) Raw ITC data, top panel and integrated heats of binding for the TolB–TolAIII complex (▪) and the corresponding data when TolB was complexed with the ColE9 T-domain ( ). Inserts, close-up views of the integrated heats for TolB–Pal + TolAIII, A and for TolB–ColE9 T-domain+TolAIII, (B). See legend to Figure 3 for conditions. Binding curves were fitted to a one-site-binding model using the Origin software. Thermodynamic data are listed in Table II.
). Inserts, close-up views of the integrated heats for TolB–Pal + TolAIII, A and for TolB–ColE9 T-domain+TolAIII, (B). See legend to Figure 3 for conditions. Binding curves were fitted to a one-site-binding model using the Origin software. Thermodynamic data are listed in Table II.

References
-
- Abergel C, Bouveret E, Claverie JM, Brown K, Rigal A, Lazdunski C, Benedetti H (1999) Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution. Structure 7: 1291–1300 - PubMed
-
- Bhatt S, Weingart C (2008) Identification of sodium chloride-regulated genes in Burkholderia cenocepacia. Curr Microbiol 56: 418–422 - PubMed
-
- Bonsor DA, Grishkovskaya I, Dodson EJ, Kleanthous C (2007) Molecular mimicry enables competitive recruitment by a natively disordered protein. J Am Chem Soc 15: 4800–4807 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

