Community-wide analysis of microbial genome sequence signatures
- PMID: 19698104
- PMCID: PMC2745766
- DOI: 10.1186/gb-2009-10-8-r85
Community-wide analysis of microbial genome sequence signatures
Abstract
Background: Analyses of DNA sequences from cultivated microorganisms have revealed genome-wide, taxa-specific nucleotide compositional characteristics, referred to as genome signatures. These signatures have far-reaching implications for understanding genome evolution and potential application in classification of metagenomic sequence fragments. However, little is known regarding the distribution of genome signatures in natural microbial communities or the extent to which environmental factors shape them.
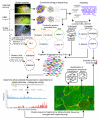
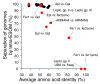
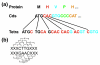
Results: We analyzed metagenomic sequence data from two acidophilic biofilm communities, including composite genomes reconstructed for nine archaea, three bacteria, and numerous associated viruses, as well as thousands of unassigned fragments from strain variants and low-abundance organisms. Genome signatures, in the form of tetranucleotide frequencies analyzed by emergent self-organizing maps, segregated sequences from all known populations sharing < 50 to 60% average amino acid identity and revealed previously unknown genomic clusters corresponding to low-abundance organisms and a putative plasmid. Signatures were pervasive genome-wide. Clusters were resolved because intra-genome differences resulting from translational selection or protein adaptation to the intracellular (pH approximately 5) versus extracellular (pH approximately 1) environment were small relative to inter-genome differences. We found that these genome signatures stem from multiple influences but are primarily manifested through codon composition, which we propose is the result of genome-specific mutational biases.
Conclusions: An important conclusion is that shared environmental pressures and interactions among coevolving organisms do not obscure genome signatures in acid mine drainage communities. Thus, genome signatures can be used to assign sequence fragments to populations, an essential prerequisite if metagenomics is to provide ecological and biochemical insights into the functioning of microbial communities.
Figures


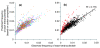
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

