Selenoprotein N is dynamically expressed during mouse development and detected early in muscle precursors
- PMID: 19698141
- PMCID: PMC2739516
- DOI: 10.1186/1471-213X-9-46
Selenoprotein N is dynamically expressed during mouse development and detected early in muscle precursors
Abstract
Background: In humans, mutations in the SEPN1 gene, encoding selenoprotein N (SelN), are involved in early onset recessive neuromuscular disorders, referred to as SEPN1-related-myopathies. The mechanisms behind these pathologies are poorly understood since the function of SelN remains elusive. However, previous results obtained in humans and more recently in zebrafish pointed to a potential role for SelN during embryogenesis. Using qRT-PCR, Western blot and whole mount in situ hybridization, we characterized in detail the spatio-temporal expression pattern of the murine Sepn1 gene during development, focusing particularly on skeletal muscles.
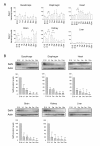
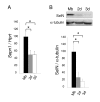
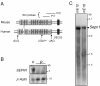
Results: In whole embryos, Sepn1 transcripts were detected as early as E5.5, with expression levels peaking at E12.5, and then strongly decreasing until birth. In isolated tissues, only mild transcriptional variations were observed during development, whereas a striking reduction of the protein expression was detected during the perinatal period. Furthermore, we demonstrated that Sepn1 is expressed early in somites and restricted to the myotome, the sub-ectodermal mesenchyme and the dorsal root ganglia at mid-gestation stages. Interestingly, Sepn1 deficiency did not alter somitogenesis in embryos, suggesting that SelN is dispensable for these processes in mouse.
Conclusion: We characterized for the first time the expression pattern of Sepn1 during mammalian embryogenesis and we demonstrated that its differential expression is most likely dependent on major post-transcriptional regulations. Overall, our data strongly suggest a potential role for selenoprotein N from mid-gestation stages to the perinatal period. Interestingly, its specific expression pattern could be related to the current hypothesis that selenoprotein N may regulate the activity of the ryanodine receptors.
Figures
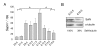


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

