Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes
- PMID: 19698979
- PMCID: PMC2750862
- DOI: 10.1016/j.cell.2009.06.049
Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes
Abstract
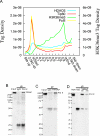
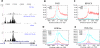
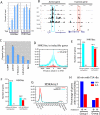
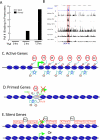
Histone acetyltransferases (HATs) and deacetylases (HDACs) function antagonistically to control histone acetylation. As acetylation is a histone mark for active transcription, HATs have been associated with active and HDACs with inactive genes. We describe here genome-wide mapping of HATs and HDACs binding on chromatin and find that both are found at active genes with acetylated histones. Our data provide evidence that HATs and HDACs are both targeted to transcribed regions of active genes by phosphorylated RNA Pol II. Furthermore, the majority of HDACs in the human genome function to reset chromatin by removing acetylation at active genes. Inactive genes that are primed by MLL-mediated histone H3K4 methylation are subject to a dynamic cycle of acetylation and deacetylation by transient HAT/HDAC binding, preventing Pol II from binding to these genes but poising them for future activation. Silent genes without any H3K4 methylation signal show no evidence of being bound by HDACs.
Figures



References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
-
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11:1587–1598. - PubMed
-
- Boyault C, Sadoul K, Pabion M, Khochbin S. HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene. 2007;26:5468–5476. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

