Chemical screen to reduce sterol accumulation in Niemann-Pick C disease cells identifies novel lysosomal acid lipase inhibitors
- PMID: 19699313
- PMCID: PMC2783675
- DOI: 10.1016/j.bbalip.2009.08.005
Chemical screen to reduce sterol accumulation in Niemann-Pick C disease cells identifies novel lysosomal acid lipase inhibitors
Abstract
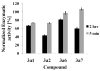
Niemann-Pick C disease (NPC) is a lysosomal storage disorder causing abnormal accumulation of unesterified free cholesterol in lysosomal storage organelles. High content phenotypic microscopy chemical screens in both human and hamster NPC-deficient cells have identified several compounds that partially revert the NPC phenotype. Cell biological and biochemical studies show that several of these molecules inhibit lysosomal acid lipase, the enzyme that hydrolyzes LDL-derived triacylglycerol and cholesteryl esters. The effects of reduced lysosomal acid lipase activity in lowering cholesterol accumulation in NPC mutant cells were verified by RNAi-mediated knockdown of lysosomal acid lipase in NPC1-deficient human fibroblasts. This work demonstrates the utility of phenotypic cellular screens as a means to identify molecular targets for altering a complex process such as intracellular cholesterol trafficking and metabolism.
Figures

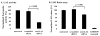




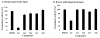
Similar articles
-
Cholesterol pathways affected by small molecules that decrease sterol levels in Niemann-Pick type C mutant cells.PLoS One. 2010 Sep 21;5(9):e12788. doi: 10.1371/journal.pone.0012788. PLoS One. 2010. PMID: 20877719 Free PMC article.
-
Automated microscopy screening for compounds that partially revert cholesterol accumulation in Niemann-Pick C cells.J Lipid Res. 2006 Feb;47(2):284-301. doi: 10.1194/jlr.M500388-JLR200. Epub 2005 Nov 15. J Lipid Res. 2006. PMID: 16288097
-
A high-content RNAi-screening assay to identify modulators of cholesterol accumulation in Niemann-Pick type C cells.Assay Drug Dev Technol. 2010 Jun;8(3):295-320. doi: 10.1089/adt.2009.0240. Assay Drug Dev Technol. 2010. PMID: 20469965
-
The potential of histone deacetylase inhibitors in Niemann - Pick type C disease.FEBS J. 2013 Dec;280(24):6367-72. doi: 10.1111/febs.12505. Epub 2013 Sep 23. FEBS J. 2013. PMID: 23992240 Free PMC article. Review.
-
NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes.J Biol Chem. 2019 Feb 1;294(5):1706-1709. doi: 10.1074/jbc.TM118.004165. J Biol Chem. 2019. PMID: 30710017 Free PMC article. Review.
Cited by
-
Identification of lysosomal lipolysis as an essential noncanonical mediator of adipocyte fasting and cold-induced lipolysis.J Clin Invest. 2025 Mar 17;135(6):e185340. doi: 10.1172/JCI185340. J Clin Invest. 2025. PMID: 40091840 Free PMC article.
-
Immunometabolic Adaptation of CD19-Targeted CAR T Cells in the Central Nervous System Microenvironment of Patients Promotes Memory Development.Cancer Res. 2024 Apr 1;84(7):1048-1064. doi: 10.1158/0008-5472.CAN-23-2299. Cancer Res. 2024. PMID: 38315779 Free PMC article.
-
δ-Tocopherol reduces lipid accumulation in Niemann-Pick type C1 and Wolman cholesterol storage disorders.J Biol Chem. 2012 Nov 16;287(47):39349-60. doi: 10.1074/jbc.M112.357707. Epub 2012 Oct 3. J Biol Chem. 2012. PMID: 23035117 Free PMC article.
-
Lysosomal acid lipase and lipophagy are constitutive negative regulators of glucose-stimulated insulin secretion from pancreatic beta cells.Diabetologia. 2014 Jan;57(1):129-39. doi: 10.1007/s00125-013-3083-x. Epub 2013 Oct 23. Diabetologia. 2014. PMID: 24149836
-
Analysis of cholesterol trafficking with fluorescent probes.Methods Cell Biol. 2012;108:367-93. doi: 10.1016/B978-0-12-386487-1.00017-1. Methods Cell Biol. 2012. PMID: 22325611 Free PMC article.
References
-
- Patterson MC, Vanier MT, Suzuki K, Morris JA, Carstea E, Neufeld EB, Blanchette-Mackie EJ, Pentchev PG. Niemann-Pick Disease TypeC: A lipid Trafficking Disorder. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Basis of Inherited Disease. Vol. 3. McGraw-Hill; New York: 2001. pp. 3611–3633.
-
- Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. - PubMed
-
- Mukherjee S, Maxfield FR. Lipid and cholesterol trafficking in NPC. Biochim Biophys Acta. 2004;1685:28–37. - PubMed
-
- Sturley SL, Patterson MC, Balch W, Liscum L. The pathophysiology and mechanisms of NP-C disease. Biochim Biophys Acta. 2004;1685:83–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

