Regulation of repair choice: Cdk1 suppresses recruitment of end joining factors at DNA breaks
- PMID: 19699692
- PMCID: PMC2748135
- DOI: 10.1016/j.dnarep.2009.07.007
Regulation of repair choice: Cdk1 suppresses recruitment of end joining factors at DNA breaks
Abstract
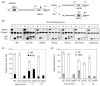
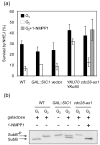
Cell cycle plays a crucial role in regulating the pathway used to repair DNA double-strand breaks (DSBs). In Saccharomyces cerevisiae, homologous recombination is primarily limited to non-G(1) cells as the formation of recombinogenic single-stranded DNA requires CDK1-dependent 5' to 3' resection of DNA ends. However, the effect of cell cycle on non-homologous end joining (NHEJ) is not yet clearly defined. Using an assay to quantitatively measure the contributions of each repair pathway to repair product formation and cellular survival after DSB induction, we found that NHEJ is most efficient at G(1), and markedly repressed at G(2). Repression of NHEJ at G(2) is achieved by efficient end resection and by the reduced association of core NHEJ proteins with DNA breaks, both of which depend on the CDK1 activity. Importantly, repression of 5' end resection by CDK1 inhibition at G(2) alone did not fully restore either physical association of Ku/Dnl4-Lif1 with DSBs or NHEJ proficiency to the level at G(1). Expression of excess Ku can partially offset the inhibition of end joining at G(2). The results suggest that regulation of Ku/Dnl4-Lif1 affinity for DNA ends may contribute to the cell cycle-dependent modulation of NHEJ efficiency.
Figures



References
-
- Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. - PubMed
-
- Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. - PubMed
-
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. - PubMed
-
- Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. - PubMed
-
- Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–826. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

