Improvement and analysis of computational methods for prediction of residual dipolar couplings
- PMID: 19700353
- PMCID: PMC2763024
- DOI: 10.1016/j.jmr.2009.07.028
Improvement and analysis of computational methods for prediction of residual dipolar couplings
Abstract
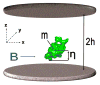
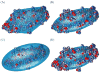
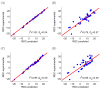
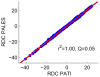
We describe a new, computationally efficient method for computing the molecular alignment tensor based on the molecular shape. The increase in speed is achieved by re-expressing the problem as one of numerical integration, rather than a simple uniform sampling (as in the PALES method), and by using a convex hull rather than a detailed representation of the surface of a molecule. This method is applicable to bicelles, PEG/hexanol, and other alignment media that can be modeled by steric restrictions introduced by a planar barrier. This method is used to further explore and compare various representations of protein shape by an equivalent ellipsoid. We also examine the accuracy of the alignment tensor and residual dipolar couplings (RDC) prediction using various ab initio methods. We separately quantify the inaccuracy in RDC prediction caused by the inaccuracy in the orientation and in the magnitude of the alignment tensor, concluding that orientation accuracy is much more important in accurate prediction of RDCs.
Figures
References
-
- Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278(5340):1111–1114. - PubMed
-
- Tycko R, Blanco FJ, Ishii Y. Alignment of biopolymers in strained gels: A new way to create detectable dipole-dipole couplings in high-resolution biomolecular NMR. Journal of the American Chemical Society. 2000;122(38):9340–9341.
-
- Sass H, Musco G, Stahl S, Wingfield P, Grzesiek S. Solution NMR of proteins within polyacrylamide gels: Diffusional properties and residual alignment by mechanical stress or embedding of oriented purple membranes. Journal of Biomolecular NMR. 2000;18(4):303–309. - PubMed
-
- Ruckert M, Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. Journal of the American Chemical Society. 2000;122(32):7793–7797.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

