Comparative transcriptomic approach to investigate differences in wine yeast physiology and metabolism during fermentation
- PMID: 19700545
- PMCID: PMC2765138
- DOI: 10.1128/AEM.01251-09
Comparative transcriptomic approach to investigate differences in wine yeast physiology and metabolism during fermentation
Abstract
Commercial wine yeast strains of the species Saccharomyces cerevisiae have been selected to satisfy many different, and sometimes highly specific, oenological requirements. As a consequence, more than 200 different strains with significantly diverging phenotypic traits are produced globally. This genetic resource has been rather neglected by the scientific community because industrial strains are less easily manipulated than the limited number of laboratory strains that have been successfully employed to investigate fundamental aspects of cellular biology. However, laboratory strains are unsuitable for the study of many phenotypes that are of significant scientific and industrial interest. Here, we investigate whether a comparative transcriptomics and phenomics approach, based on the analysis of five phenotypically diverging industrial wine yeast strains, can provide insights into the molecular networks that are responsible for the expression of such phenotypes. For this purpose, some oenologically relevant phenotypes, including resistance to various stresses, cell wall properties, and metabolite production of these strains were evaluated and aligned with transcriptomic data collected during alcoholic fermentation. The data reveal significant differences in gene regulation between the five strains. While the genetic complexity underlying the various successive stress responses in a dynamic system such as wine fermentation reveals the limits of the approach, many of the relevant differences in gene expression can be linked to specific phenotypic differences between the strains. This is, in particular, the case for many aspects of metabolic regulation. The comparative approach therefore opens new possibilities to investigate complex phenotypic traits on a molecular level.
Figures
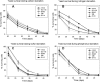

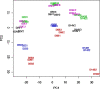


Similar articles
-
Physical cell-cell contact elicits specific transcriptomic responses in wine yeast species.Microbiol Spectr. 2024 Aug 6;12(8):e0057223. doi: 10.1128/spectrum.00572-23. Epub 2024 Jul 16. Microbiol Spectr. 2024. PMID: 39012115 Free PMC article.
-
Linking gene regulation and the exo-metabolome: a comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast.BMC Genomics. 2008 Nov 7;9:530. doi: 10.1186/1471-2164-9-530. BMC Genomics. 2008. PMID: 18990252 Free PMC article.
-
FLO gene-dependent phenotypes in industrial wine yeast strains.Appl Microbiol Biotechnol. 2010 Apr;86(3):931-45. doi: 10.1007/s00253-009-2381-1. Epub 2009 Dec 16. Appl Microbiol Biotechnol. 2010. PMID: 20013339
-
Biotechnological impact of stress response on wine yeast.Lett Appl Microbiol. 2017 Feb;64(2):103-110. doi: 10.1111/lam.12677. Epub 2016 Nov 21. Lett Appl Microbiol. 2017. PMID: 27714822 Review.
-
From yeast genetics to biotechnology.Acta Microbiol Immunol Hung. 2002;49(4):483-91. doi: 10.1556/AMicr.49.2002.4.6. Acta Microbiol Immunol Hung. 2002. PMID: 12512257 Review.
Cited by
-
Genomic expression program of Saccharomyces cerevisiae along a mixed-culture wine fermentation with Hanseniaspora guilliermondii.Microb Cell Fact. 2015 Aug 28;14:124. doi: 10.1186/s12934-015-0318-1. Microb Cell Fact. 2015. PMID: 26314747 Free PMC article.
-
Adjustment of trehalose metabolism in wine Saccharomyces cerevisiae strains to modify ethanol yields.Appl Environ Microbiol. 2013 Sep;79(17):5197-207. doi: 10.1128/AEM.00964-13. Epub 2013 Jun 21. Appl Environ Microbiol. 2013. PMID: 23793638 Free PMC article.
-
Genome-wide study of the adaptation of Saccharomyces cerevisiae to the early stages of wine fermentation.PLoS One. 2013 Sep 5;8(9):e74086. doi: 10.1371/journal.pone.0074086. eCollection 2013. PLoS One. 2013. PMID: 24040173 Free PMC article.
-
Effects of Active Dry Yeast Supplementation in In Vitro and In Vivo Nutrient Digestibility, Rumen Fermentation, and Bacterial Community.Animals (Basel). 2024 Oct 9;14(19):2916. doi: 10.3390/ani14192916. Animals (Basel). 2024. PMID: 39409865 Free PMC article.
-
Comparative transcriptomic and proteomic profiling of industrial wine yeast strains.Appl Environ Microbiol. 2010 Jun;76(12):3911-23. doi: 10.1128/AEM.00586-10. Epub 2010 Apr 23. Appl Environ Microbiol. 2010. PMID: 20418425 Free PMC article.
References
-
- Abbott, D. A., T. A. Knijnenburg, L. M. de Poorter, M. J. Reinders, J. T. Pronk, and A. J. van Maris. 2007. Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae. FEMS Yeast Res. 7:819-833. - PubMed
-
- Attfield, P. V. 1997. Stress tolerance: the key to effective strains of baker's yeast. Nat. Biotechnol. 15:1351-1357. - PubMed
-
- Bauer, F. F., and I. S. Pretorius. 2000. Yeast stress response and fermentation efficiency: how to survive the making of wine—a review. S. Afr. J. Enol. 21:27-51.
-
- Beltran, G., M. Novo, V. Leberre, S. Sokol, D. Labourdette, J. M. Guillamon, A. Mas, J. François, and N. Rozes. 2006. Integration of transcriptomic and metabolic analyses for understanding the global responses of low-temperature winemaking fermentations. FEMS Yeast Res. 6:1167-1183. - PubMed
-
- Bely, L., J. Sablayrolles, and P. Barre. 1990. Description of alcoholic fermentation kinetics: its variability and significance. Am. J. Enol. Viticult. 41:319-324.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

