Recovery of nonpathogenic mutant bacteria from tumors caused by several Agrobacterium tumefaciens strains: a frequent event?
- PMID: 19700547
- PMCID: PMC2765143
- DOI: 10.1128/AEM.01867-08
Recovery of nonpathogenic mutant bacteria from tumors caused by several Agrobacterium tumefaciens strains: a frequent event?
Abstract
We have evaluated the interaction that bacterial genotypes and plant hosts have with the loss of pathogenicity in tumors, using seven Agrobacterium tumefaciens strains inoculated on 12 herbaceous and woody hosts. We performed a screening of the agrobacteria present inside the tumors, looking for nonpathogenic strains, and found a high variability of those strains in this niche. To verify the origin of the putative nonpathogenic mutant bacteria, we applied an efficient, reproducible, and specific randomly amplified polymorphic DNA analysis method. In contrast with previous studies, we recovered a very small percentage (0.01%) of nonpathogenic strains that can be considered true mutants. Of 5,419 agrobacterial isolates examined, 662 were nonpathogenic in tomato, although only 7 (from pepper and tomato tumors induced by two A. tumefaciens strains) could be considered to derive from the inoculated strain. Six mutants were affected in the transferred DNA (T-DNA) region; one of them contained IS426 inserted into the iaaM gene, whereas the whole T-DNA region was apparently deleted in three other mutants, and the virulence of the remaining two mutants was fully restored with the T-DNA genes as well. The plasmid profile was altered in six of the mutants, with changes in the size of the Ti plasmid or other plasmids and/or the acquisition of new plasmids. Our results also suggest that the frequent occurrence of nonpathogenic clones in the tumors is probably due to the preferential growth of nonpathogenic agrobacteria, of either endophytic or environmental origin, but different from the bacterial strain inducing the tumor.
Figures

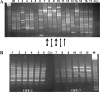


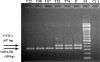


Similar articles
-
Genetic analysis of nonpathogenic Agrobacterium tumefaciens mutants arising in crown gall tumors.J Bacteriol. 1995 Jul;177(13):3752-7. doi: 10.1128/jb.177.13.3752-3757.1995. J Bacteriol. 1995. PMID: 7601840 Free PMC article.
-
pSa causes oncogenic suppression of Agrobacterium by inhibiting VirE2 protein export.J Bacteriol. 1999 Jan;181(1):186-96. doi: 10.1128/JB.181.1.186-196.1999. J Bacteriol. 1999. PMID: 9864329 Free PMC article.
-
Complete sequence of the tumor-inducing plasmid pTiChry5 from the hypervirulent Agrobacterium tumefaciens strain Chry5.Plasmid. 2018 Mar-May;96-97:1-6. doi: 10.1016/j.plasmid.2018.02.001. Epub 2018 Feb 7. Plasmid. 2018. PMID: 29427647
-
[Opine biosynthesis and catabolism genes of Agrobacterium tumefaciens and Agrobacterium rhizogenes].Genetika. 2015 Feb;51(2):137-46. Genetika. 2015. PMID: 25966579 Review. Russian.
-
The Agrobacterium Ti Plasmids.Microbiol Spectr. 2014 Dec;2(6):10.1128/microbiolspec.PLAS-0010-2013. doi: 10.1128/microbiolspec.PLAS-0010-2013. Microbiol Spectr. 2014. PMID: 25593788 Free PMC article. Review.
Cited by
-
Endophytic bacterial diversity in banana 'Prata Anã' (Musa spp.) roots.Genet Mol Biol. 2013 Jul;36(2):252-64. doi: 10.1590/S1415-47572013000200016. Epub 2013 Jun 22. Genet Mol Biol. 2013. PMID: 23885208 Free PMC article.
-
Quorum-quenching limits quorum-sensing exploitation by signal-negative invaders.Sci Rep. 2017 Jan 5;7:40126. doi: 10.1038/srep40126. Sci Rep. 2017. PMID: 28054641 Free PMC article.
-
Ecological and evolutionary dynamics of a model facultative pathogen: Agrobacterium and crown gall disease of plants.Environ Microbiol. 2018 Jan;20(1):16-29. doi: 10.1111/1462-2920.13976. Epub 2017 Dec 4. Environ Microbiol. 2018. PMID: 29105274 Free PMC article. Review.
-
Ecological dynamics and complex interactions of Agrobacterium megaplasmids.Front Plant Sci. 2014 Nov 14;5:635. doi: 10.3389/fpls.2014.00635. eCollection 2014. Front Plant Sci. 2014. PMID: 25452760 Free PMC article. Review.
-
Plant GABA:proline ratio modulates dissemination of the virulence Ti plasmid within the Agrobacterium tumefaciens hosted population.Plant Signal Behav. 2016 May 3;11(5):e1178440. doi: 10.1080/15592324.2016.1178440. Plant Signal Behav. 2016. PMID: 27110651 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Anderson, A. R., and L. W. Moore. 1979. Host specificity in the genus Agrobacterium. Phytopathology 69:320-323.
-
- Bélanger, C., M. L. Canfield, L. W. Moore, and P. Dion. 1993. Detection of avirulent mutants of Agrobacterium tumefaciens in crown-gall tumors produced in vitro. p. 97-101. In E. W. Nester and D. P. S. Verma (ed.), Advances in molecular genetics of plant microbe interactions, vol. 2. Kluwer Academic Publishers, Dordrecht, The Netherlands.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

