The transcription factor homolog CTF1 regulates {beta}-oxidation in Candida albicans
- PMID: 19700635
- PMCID: PMC2756860
- DOI: 10.1128/EC.00206-09
The transcription factor homolog CTF1 regulates {beta}-oxidation in Candida albicans
Abstract
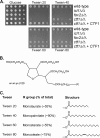
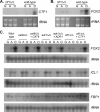
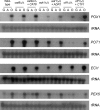
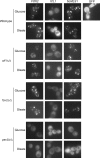
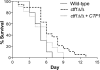
Carbon starvation is one of the many stresses to which microbial pathogens are subjected while in the host. Pathways necessary for the utilization of alternative carbon sources, such as gluconeogenesis, the glyoxylate cycle, and beta-oxidation of fatty acids, have been shown to be required for full virulence in several systems, including the fungal pathogen Candida albicans. We have investigated the regulatory network governing alternative carbon metabolism in this organism through characterization of transcriptional regulators identified based on the model fungi, Saccharomyces cerevisiae and Aspergillus nidulans. C. albicans has homologs of the ScCAT8/AnFacB and ScADR1/AnAmdX transcription factors that regulate induction of genes encoding the proteins of gluconeogenesis, the glyoxylate cycle, and ethanol utilization. Surprisingly, C. albicans mutants lacking CAT8 or ADR1 have no apparent phenotypes and do not regulate genes for key enzymes of these pathways. Fatty acid degradation and peroxisomal biogenesis are controlled by nonhomologous regulators, OAF1/PIP2 in S. cerevisiae and FarA/FarB in A. nidulans; C. albicans is missing OAF1 and PIP2 and, instead, has a single homolog of the Far proteins, CTF1. We have shown that CTF1 is required for growth on lipids and for expression of genes necessary for beta-oxidation, such as FOX2. ctf1Delta/ctf1Delta (ctf1Delta/Delta) strains do not, however, show the pleiotropic phenotypes observed for fox2Delta/Delta mutants. The ctf1Delta/Delta mutant confers a mild attenuation in virulence, like the fox2Delta/Delta mutant. Thus, phenotypic and genotypic observations highlight important differences in the regulatory network for alternative carbon metabolism in C. albicans compared to the paradigms developed in other model fungi.
Figures



Similar articles
-
Peroxisomal fatty acid beta-oxidation is not essential for virulence of Candida albicans.Eukaryot Cell. 2006 Nov;5(11):1847-56. doi: 10.1128/EC.00093-06. Epub 2006 Sep 8. Eukaryot Cell. 2006. PMID: 16963628 Free PMC article.
-
Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes.Eukaryot Cell. 2007 Feb;6(2):280-90. doi: 10.1128/EC.00372-06. Epub 2006 Dec 8. Eukaryot Cell. 2007. PMID: 17158734 Free PMC article.
-
The activity of the glyoxylate cycle in peroxisomes of Candida albicans depends on a functional beta-oxidation pathway: evidence for reduced metabolite transport across the peroxisomal membrane.Microbiology (Reading). 2008 Oct;154(Pt 10):3061-3072. doi: 10.1099/mic.0.2008/020289-0. Microbiology (Reading). 2008. PMID: 18832312
-
The glyoxylate cycle and alternative carbon metabolism as metabolic adaptation strategies of Candida glabrata: perspectives from Candida albicans and Saccharomyces cerevisiae.J Biomed Sci. 2019 Jul 13;26(1):52. doi: 10.1186/s12929-019-0546-5. J Biomed Sci. 2019. PMID: 31301737 Free PMC article. Review.
-
Peroxisome biogenesis in Saccharomyces cerevisiae.Antonie Van Leeuwenhoek. 1992 Aug;62(1-2):63-78. doi: 10.1007/BF00584463. Antonie Van Leeuwenhoek. 1992. PMID: 1444337 Review.
Cited by
-
A potent plant-derived antifungal acetylenic acid mediates its activity by interfering with fatty acid homeostasis.Antimicrob Agents Chemother. 2012 Jun;56(6):2894-907. doi: 10.1128/AAC.05663-11. Epub 2012 Mar 19. Antimicrob Agents Chemother. 2012. PMID: 22430960 Free PMC article.
-
Role of carnitine acetyltransferases in acetyl coenzyme A metabolism in Aspergillus nidulans.Eukaryot Cell. 2011 Apr;10(4):547-55. doi: 10.1128/EC.00295-10. Epub 2011 Feb 4. Eukaryot Cell. 2011. PMID: 21296915 Free PMC article.
-
Two homologs of the Cat8 transcription factor are involved in the regulation of ethanol utilization in Komagataella phaffii.Curr Genet. 2021 Aug;67(4):641-661. doi: 10.1007/s00294-021-01165-4. Epub 2021 Mar 16. Curr Genet. 2021. PMID: 33725138 Free PMC article.
-
Adaptation of Candida albicans to specific host environments by gain-of-function mutations in transcription factors.PLoS Pathog. 2024 Nov 4;20(11):e1012643. doi: 10.1371/journal.ppat.1012643. eCollection 2024 Nov. PLoS Pathog. 2024. PMID: 39495716 Free PMC article. Review.
-
Manipulation of Host Diet To Reduce Gastrointestinal Colonization by the Opportunistic Pathogen Candida albicans.mSphere. 2015 Nov 18;1(1):e00020-15. doi: 10.1128/mSphere.00020-15. eCollection 2016 Jan-Feb. mSphere. 2015. PMID: 27303684 Free PMC article.
References
-
- Ausubel, F. M., B. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley & Sons, Edison, NJ.
-
- Barelle, C. J., C. L. Manson, D. M. MacCallum, F. C. Odds, N. A. Gow, and A. J. Brown. 2004. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast 21:333-340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

