Transgenic CHD1L expression in mouse induces spontaneous tumors
- PMID: 19701453
- PMCID: PMC2726430
- DOI: 10.1371/journal.pone.0006727
Transgenic CHD1L expression in mouse induces spontaneous tumors
Abstract
Background: Amplification of 1q21 is the most frequent genetic alteration in hepatocellular carcinoma (HCC), which was detected in 58-78% of primary HCC cases by comparative genomic hybridization (CGH). Using chromosome microdissection/hybrid selection approach we recently isolated a candidate oncogene CHD1L from 1q21 region. Our previous study has demonstrated that CHD1L had strong oncogenic ability, which could be effectively suppressed by siRNA against CHD1L. The molecular mechanism of CHD1L in tumorigenesis has been associated with its role in promoting cell proliferation.
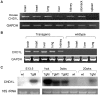
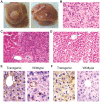
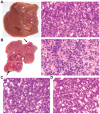
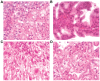
Methodology/principal findings: To further investigate the in vivo oncogenic role of CHD1L, CHD1L ubiquitous-expression transgenic mouse model was generated. Spontaneous tumor formations were found in 10/41 (24.4%) transgenic mice, including 4 HCCs, but not in their 39 wild-type littermates. In addition, alcohol intoxication was used to induce hepatocyte pathological lesions and results found that overexpression of CHD1L in hepatocytes could promote tumor susceptibility in CHD1L-transgenic mice. To address the mechanism of CHD1L in promoting cell proliferation, DNA content between CHD1L-transgenic and wildtype mouse embryo fibroblasts (MEFs) was compared by flow cytometry. Flow cytometry results found that CHD1L could facilitate DNA synthesis and G1/S transition through the up-regulation of Cyclin A, Cyclin D1, Cyclin E, CDK2, and CDK4, and down-regulation of Rb, p27(Kip1), and p53.
Conclusion/significance: Taken together, our data strongly support that CHD1L is a novel oncogene and plays an important role in HCC pathogenesis.
Conflict of interest statement
Figures


References
-
- Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. - PubMed
-
- El-Serag HB, Mason AC, Key C. Trends in survival of patients with hepatocellular carcinoma between 1977 and 1996 in the United States. Hepatology. 2001;33:62–65. - PubMed
-
- Montalto G, Gervello M, Giannitrapani L, Dantona F, Terranova A, et al. Epidemiology, risk factors, and natural history of hepatocellular carcinoma. Ann NY Acad Sci. 2002;963:13–20. - PubMed
-
- Marchio A, Meddeb M, Pineau P, Danglot G, Tiollais P, et al. Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer. 1997;18:59–65. - PubMed
-
- Kusano N, Shiraishi K, Kubo K, Oga A, Okita K, et al. Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology. 1999;29:1858–1862. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

