Designed zinc finger protein interacting with the HIV-1 integrase recognition sequence at 2-LTR-circle junctions
- PMID: 19701937
- PMCID: PMC2788277
- DOI: 10.1002/pro.233
Designed zinc finger protein interacting with the HIV-1 integrase recognition sequence at 2-LTR-circle junctions
Abstract
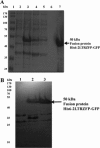
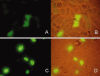
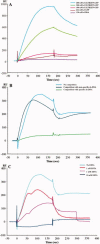
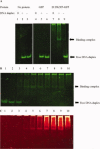
Integration of HIV-1 cDNA into the host genome is a crucial step for viral propagation. Two nucleotides, cytosine and adenine (CA), conserved at the 3' end of the viral cDNA genome, are cleaved by the viral integrase (IN) enzyme. As IN plays a crucial role in the early stages of the HIV-1 life cycle, substrate blockage of IN is an attractive strategy for therapeutic interference. In this study, we used the 2-LTR-circle junctions of HIV-1 DNA as a model to design zinc finger protein (ZFP) targeting at the end terminal portion of HIV-1 LTR. A six-contiguous ZFP, namely 2LTRZFP was designed using zinc finger tools. The designed motif was expressed and purified from E. coli to determine its binding properties. Surface plasmon resonance (SPR) was used to determine the binding affinity of 2LTRZFP to its target DNA. The level of dissociation constant (K(d)) was 12.0 nM. The competitive SPR confirmed that 2LTRZFP specifically interacted with its target DNA. The qualitative binding activity was subsequently determined by EMSA and demonstrated the aforementioned correlation. In addition, molecular modeling and binding energy analyses were carried out to provide structural insight into the binding of 2LTRZFP to the specific and nonspecific DNA target. It is suggested that hydrogen-bonding interactions play a key role in the DNA recognition mechanisms of the designed ZFP. Our study suggested an alternative HIV therapeutic strategy using ZFP interference of the HIV integration process.
Figures



Similar articles
-
Zinc finger protein designed to target 2-long terminal repeat junctions interferes with human immunodeficiency virus integration.Hum Gene Ther. 2012 Sep;23(9):932-42. doi: 10.1089/hum.2011.124. Epub 2012 May 8. Hum Gene Ther. 2012. PMID: 22429108 Free PMC article.
-
Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites.J Virol. 2004 Feb;78(3):1301-13. doi: 10.1128/jvi.78.3.1301-1313.2004. J Virol. 2004. PMID: 14722285 Free PMC article.
-
2LTRZFP Interacts Specifically to HIV-1 DNA without Off-Target Effects as Determined by Biolayer Interferometry.Biosensors (Basel). 2021 Mar 8;11(3):76. doi: 10.3390/bios11030076. Biosensors (Basel). 2021. PMID: 33800287 Free PMC article.
-
Site-specific integration of retroviral DNA in human cells using fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc-finger protein E2C.Methods. 2009 Apr;47(4):269-76. doi: 10.1016/j.ymeth.2009.01.001. Epub 2009 Jan 30. Methods. 2009. PMID: 19186211 Free PMC article. Review.
-
Characterization and structural analysis of HIV-1 integrase conservation.AIDS Rev. 2009 Jan-Mar;11(1):17-29. AIDS Rev. 2009. PMID: 19290031 Review.
Cited by
-
Molecular dynamics study on the strengthening behavior of Delta and Omicron SARS-CoV-2 spike RBD improved receptor-binding affinity.PLoS One. 2022 Nov 17;17(11):e0277745. doi: 10.1371/journal.pone.0277745. eCollection 2022. PLoS One. 2022. PMID: 36395151 Free PMC article.
-
Zinc finger protein designed to target 2-long terminal repeat junctions interferes with human immunodeficiency virus integration.Hum Gene Ther. 2012 Sep;23(9):932-42. doi: 10.1089/hum.2011.124. Epub 2012 May 8. Hum Gene Ther. 2012. PMID: 22429108 Free PMC article.
-
An integrative approach for a network based meta-analysis of viral RNAi screens.Algorithms Mol Biol. 2015 Feb 13;10:6. doi: 10.1186/s13015-015-0035-7. eCollection 2015. Algorithms Mol Biol. 2015. PMID: 25691914 Free PMC article.
-
Modulated expression of the HIV-1 2LTR zinc finger efficiently interferes with the HIV integration process.Biosci Rep. 2018 Sep 7;38(5):BSR20181109. doi: 10.1042/BSR20181109. Print 2018 Oct 31. Biosci Rep. 2018. PMID: 30068696 Free PMC article.
-
Performance of Affinity-Improved DARPin Targeting HIV Capsid Domain in Interference of Viral Progeny Production.Biomolecules. 2021 Sep 30;11(10):1437. doi: 10.3390/biom11101437. Biomolecules. 2021. PMID: 34680070 Free PMC article.
References
-
- Mann JM, Chin J, Piot P, Quinn T. The international epidemiology of AIDS. Sci Am. 1988;259:82–89. - PubMed
-
- UNAIDS/World Health Organization (2008) Report on the Global HIV/AIDS Epidemic. August, Mexico City.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

