Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II
- PMID: 19703992
- PMCID: PMC2772563
- DOI: 10.1128/MCB.00924-09
Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II
Abstract
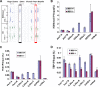
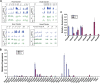
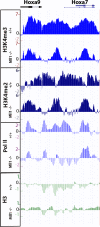
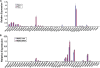
A common landmark of activated genes is the presence of trimethylation on lysine 4 of histone H3 (H3K4) at promoter regions. Set1/COMPASS was the founding member and is the only H3K4 methylase in Saccharomyces cerevisiae; however, in mammals, at least six H3K4 methylases, Set1A and Set1B and MLL1 to MLL4, are found in COMPASS-like complexes capable of methylating H3K4. To gain further insight into the different roles and functional targets for the H3K4 methylases, we have undertaken a genome-wide analysis of H3K4 methylation patterns in wild-type Mll1(+/+) and Mll1(-)(/)(-) mouse embryonic fibroblasts (MEFs). We found that Mll1 is required for the H3K4 trimethylation of less than 5% of promoters carrying this modification. Many of these genes, which include developmental regulators such as Hox genes, show decreased levels of RNA polymerase II recruitment and expression concomitant with the loss of H3K4 methylation. Although Mll1 is only required for the methylation of a subset of Hox genes, menin, a component of the Mll1 and Mll2 complexes, is required for the overwhelming majority of H3K4 methylation at Hox loci. However, the loss of MLL3/MLL4 and/or the Set1 complexes has little to no effect on the H3K4 methylation of Hox loci or their expression levels in these MEFs. Together these data provide insight into the redundancy and specialization of COMPASS-like complexes in mammals and provide evidence for a possible role for Mll1-mediated H3K4 methylation in the regulation of transcriptional initiation.
Figures




References
-
- Allis, C. D., S. L. Berger, J. Cote, S. Dent, T. Jenuwien, T. Kouzarides, L. Pillus, D. Reinberg, Y. Shi, R. Shiekhattar, A. Shilatifard, J. Workman, and Y. Zhang. 2007. New nomenclature for chromatin-modifying enzymes. Cell 131:633-636. - PubMed
-
- Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20:5695-5707. - PubMed
-
- Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169-181. - PubMed
-
- Charité, J., W. de Graaff, D. Consten, M. J. Reijnen, J. Korving, and J. Deschamps. 1998. Transducing positional information to the Hox genes: critical interaction of cdx gene products with position-sensitive regulatory elements. Development 125:4349-4358. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
