Functional interaction between the Fanconi Anemia D2 protein and proliferating cell nuclear antigen (PCNA) via a conserved putative PCNA interaction motif
- PMID: 19704162
- PMCID: PMC2781439
- DOI: 10.1074/jbc.M109.016352
Functional interaction between the Fanconi Anemia D2 protein and proliferating cell nuclear antigen (PCNA) via a conserved putative PCNA interaction motif
Abstract
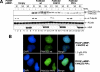
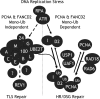
Fanconi Anemia (FA) is a rare recessive disease characterized by congenital abnormalities, bone marrow failure, and cancer susceptibility. The FA proteins and the familial breast cancer susceptibility gene products, BRCA1 and FANCD1/BRCA2, function cooperatively in the FA-BRCA pathway to repair damaged DNA and to prevent cellular transformation. Activation of this pathway occurs via the mono-ubiquitination of the FANCD2 protein, targeting it to nuclear foci where it co-localizes with FANCD1/BRCA2, RAD51, and PCNA. The regulation of the mono-ubiquitination of FANCD2, as well as its function in DNA repair remain poorly understood. In this study, we have further characterized the interaction between the FANCD2 and PCNA proteins. We have identified a highly conserved, putative FANCD2 PCNA interaction motif (PIP-box), and demonstrate that mutation of this motif disrupts FANCD2-PCNA binding and precludes the mono-ubiquitination of FANCD2. Consequently, the FANCD2 PIP-box mutant protein fails to correct the mitomycin C hypersensitivity of FA-D2 patient cells. Our results suggest that PCNA may function as a molecular platform to facilitate the mono-ubiquitination of FANCD2 and activation of the FA-BRCA pathway.
Figures





References
-
- Taniguchi T., D'Andrea A. D. (2006) Blood 107, 4223–4233 - PubMed
-
- Fanconi Anemia: Standards for Clinical Care, 2nd Ed., Fanconi Anemia Research Fund, Inc., Eugene, OR
-
- Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M. S., Timmers C., Hejna J., Grompe M., D'Andrea A. D. (2001) Mol. Cell. 7, 249–262 - PubMed
-
- Howlett N. G., Taniguchi T., Durkin S. G., D'Andrea A. D., Glover T. W. (2005) Hum. Mol. Genet. 14, 693–701 - PubMed
-
- Wang W. (2007) Nat. Rev. Genet. 8, 735–748 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

