Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival
- PMID: 19706168
- PMCID: PMC2753634
- DOI: 10.1186/1471-2407-9-298
Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival
Abstract
Background: The transcription factor GLI1, a member of the GLI subfamily of Krüppel-like zinc finger proteins is involved in signal transduction within the hedgehog pathway. Aberrant hedgehog signalling has been implicated in the development of different human tumour entities such as colon and lung cancer and increased GLI1 expression has been found in these tumour entities as well. In this study we questioned whether GLI1 expression might also be important in human breast cancer development. Furthermore we correlated GLI1 expression with histopathological and clinical data to evaluate whether GLI1 could represent a new prognostic marker in breast cancer treatment.
Methods: Applying semiquantitative realtime PCR analysis and immunohistochemistry (IHC) GLI1 expression was analysed in human invasive breast carcinomas (n = 229) in comparison to normal human breast tissues (n = 58). GLI1 mRNA expression was furthermore analysed in a set of normal (n = 3) and tumourous breast cell lines (n = 8). IHC data were statistically interpreted using SPSS version 14.0.
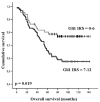
Results: Initial analysis of GLI1 mRNA expression in a small cohort of (n = 5) human matched normal and tumourous breast tissues showed first tendency towards GLI1 overexpression in human breast cancers. However only a small sample number was included into these analyses and values for GLI1 overexpression were statistically not significant (P = 0.251, two-tailed Mann-Whitney U-test). On protein level, nuclear GLI1 expression in breast cancer cells was clearly more abundant than in normal breast epithelial cells (P = 0.008, two-tailed Mann-Whitney U-test) and increased expression of GLI1 protein in breast tumours significantly correlated with unfavourable overall survival (P = 0.019), but also with higher tumour stage (P < 0.001) and an increased number of tumour-positive axillar lymph nodes (P = 0.027). Interestingly, a highly significant correlation was found between GLI1 expression and the expression of SHH, a central upstream molecule of the hedgehog pathway that was previously analysed on the same tissue microarray.
Conclusion: Our study presents a systematic expression analysis of GLI1 in human breast cancer. Elevated levels of GLI1 protein in human breast cancer are associated with unfavourable prognosis and progressive stages of disease. Thus GLI1 protein expression measured e.g. by an IHC based scoring system might have an implication in future multi-marker panels for human breast cancer prognosis or molecular sub typing. The highly significant correlation between SHH and GLI1 expression characterises GLI1 as a potential functional downstream target of the hedgehog signalling pathway in human breast cancer as well. Furthermore, our study indicates that altered hedgehog signalling may represent a key disease pathway in the progression of human breast cancer.
Figures

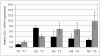

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

