Claudin-16 and claudin-19 interaction is required for their assembly into tight junctions and for renal reabsorption of magnesium
- PMID: 19706394
- PMCID: PMC2741254
- DOI: 10.1073/pnas.0907724106
Claudin-16 and claudin-19 interaction is required for their assembly into tight junctions and for renal reabsorption of magnesium
Abstract
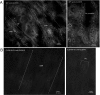
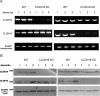
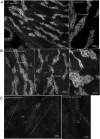
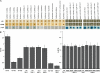
Claudins are tight junction integral membrane proteins that are key regulators of the paracellular pathway. Defects in claudin-16 (CLDN16) and CLDN19 function result in the inherited human renal disorder familial hypomagnesemia with hypercalciuria and nephrocalcinosis (FHHNC). Previous studies showed that siRNA knockdown of CLDN16 in mice results in a mouse model for FHHNC. Here, we show that CLDN19-siRNA mice also developed the FHHNC symptoms of chronic renal wasting of magnesium and calcium together with defective renal salt handling. siRNA knockdown of CLDN19 caused a loss of CLDN16 from tight junctions in the thick ascending limb (TAL) without a decrease in CLDN16 expression level, whereas siRNA knockdown of CLDN16 produced a similar effect on CLDN19. In both mouse lines, CLDN10, CLDN18, occludin, and ZO-1, normal constituents of TAL tight junctions, remained correctly localized. CLDN16- and CLDN19-depleted tight junctions had normal barrier function but defective ion selectivity. These data, together with yeast two-hybrid binding studies, indicate that a heteromeric CLDN16 and CLDN19 interaction was required for assembling them into the tight junction structure and generating cation-selective paracellular channels.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

