A 3D structure model of the melibiose permease of Escherichia coli represents a distinctive fold for Na+ symporters
- PMID: 19706416
- PMCID: PMC2729278
- DOI: 10.1073/pnas.0905516106
A 3D structure model of the melibiose permease of Escherichia coli represents a distinctive fold for Na+ symporters
Abstract
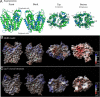
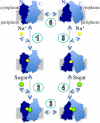
The melibiose permease of Escherichia coli (MelB) catalyzes the coupled stoichiometric symport of a galactoside with a cation (either Na(+), Li(+), or H(+)), using free energy from the downhill translocation of one cosubstrate to catalyze the accumulation of the other. Here, we present a 3D structure model of MelB threaded through a crystal structure of the lactose permease of E. coli (LacY), manually adjusted, and energetically minimized. The model contains 442 consecutive residues ( approximately 94% of the polypeptide), including all 12 transmembrane helices and connecting loops, with no steric clashes and superimposes well with the template structure. The electrostatic surface potential calculated from the model is typical for a membrane protein and exhibits a characteristic ring of positive charges around the periphery of the cytoplasmic side. The 3D model indicates that MelB consists of two pseudosymmetrical 6-helix bundles lining an internal hydrophilic cavity, which faces the cytoplasmic side of the membrane. Both sugar and cation binding sites are proposed to lie within the internal cavity. The model is consistent with numerous previous mutational, biochemical/biophysical characterizations as well as low-resolution structural data. Thus, an alternating access mechanism with sequential binding is discussed. The proposed overall fold of MelB is different from the available crystal structures of other Na(+)-coupled transporters, suggesting a distinctive fold for Na(+) symporters.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


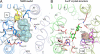
References
-
- Yazyu H, et al. Nucleotide sequence of the melB gene and characteristics of deduced amino acid sequence of the melibiose carrier in Escherichia coli. J Biol Chem. 1984;259:4320–4326. - PubMed
-
- Saier MH., Jr Families of transmembrane sugar transport proteins. Mol Microbiol. 2000;35:699–710. - PubMed
-
- Tokuda H, Kaback HR. Sodium-dependent methyl 1-thio-β-d-galactopyranoside transport in membrane vesicles isolated from Salmonella typhimurium. Biochemistry. 1977;16:2130–2136. - PubMed
-
- Tsuchiya T, Raven J, Wilson TH. Co-transport of Na+ and methul-beta-d-thiogalactopyranoside mediated by the melibiose transport system of Escherichia coli. Biochem Biophys Res Commun. 1977;76:26–31. - PubMed
-
- Bassilana M, Pourcher T, Leblanc G. Facilitated diffusion properties of melibiose permease in Escherichia coli membrane vesicles. Release of co-substrates is rate limiting for permease cycling. J Biol Chem. 1987;262:16865–16870. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

