The human IgM pentamer is a mushroom-shaped molecule with a flexural bias
- PMID: 19706439
- PMCID: PMC2736442
- DOI: 10.1073/pnas.0903805106
The human IgM pentamer is a mushroom-shaped molecule with a flexural bias
Abstract
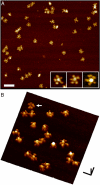
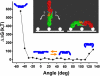
The textbook planar model of pentameric IgM, a potent activator of complement C1q, is based upon the crystallographic structure of IgG. Although widely accepted, key predictions of this model have not yet been directly confirmed, which is particularly important since IgG lacks a major Ig fold domain in its Fc region that is present in IgM. Here, we construct a homology-based structural model of the IgM pentamer using the recently obtained crystallographic structure of IgE Fc, which has this additional Ig domain, under the constraint that all of the cysteine residues known to form disulfide bridges both within each monomer and between monomers are bonded together. In contrast to the planar model, this model predicts a non-planar, mushroom-shaped complex, with the central portion formed by the C-terminal domains protruding out of the plane formed by the Fab domains. This unexpected conformation of IgM is, however, directly confirmed by cryo-atomic force microscopy of individual human IgM molecules. Further analysis of this model with free energy calculations of out-of-plane Fab domain rotations reveals a pronounced asymmetry favoring flexions toward the central protrusion. This bias, together with polyvalent attachment to cell surface antigen, would ensure that the IgM pentamer is oriented on the cell membrane with its C1q binding sites fully exposed to the solution, and thus provides a mechanistic explanation for the first steps of C1q activation by IgM.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Ochsenbein AF, et al. Control of early viral and bacterial distribution and disease by natural antibodies. Science. 1999;286:2156–2159. - PubMed
-
- Vollmers HP, Brandlein S. Natural IgM antibodies: The orphaned molecules in immune surveillance. Adv Drug Deliv Rev. 2006;58:755–765. - PubMed
-
- Beutner U, et al. Neoadjuvant therapy of gastric cancer with the human monoclonal IgM antibody SC-1: Impact on the immune system. Oncol Rep. 2008;19:761–769. - PubMed
-
- Illert B, et al. Human antibody SC-1 reduces disseminated tumor cells in nude mice with human gastric cancer. Oncol Rep. 2005;13:765–770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

