Mapping accessible chromatin regions using Sono-Seq
- PMID: 19706456
- PMCID: PMC2736440
- DOI: 10.1073/pnas.0905443106
Mapping accessible chromatin regions using Sono-Seq
Abstract
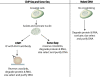
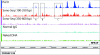
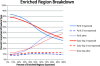
Disruptions in local chromatin structure often indicate features of biological interest such as regulatory regions. We find that sonication of cross-linked chromatin, when combined with a size-selection step and massively parallel short-read sequencing, can be used as a method (Sono-Seq) to map locations of high chromatin accessibility in promoter regions. Sono-Seq sites frequently correspond to actively transcribed promoter regions, as evidenced by their co-association with RNA Polymerase II ChIP regions, transcription start sites, histone H3 lysine 4 trimethylation (H3K4me3) marks, and CpG islands; signals over other sites, such as those bound by the CTCF insulator, are also observed. The pattern of breakage by Sono-Seq overlaps with, but is distinct from, that observed for FAIRE and DNase I hypersensitive sites. Our results demonstrate that Sono-Seq can be a useful and simple method by which to map many local alterations in chromatin structure. Furthermore, our results provide insights into the mapping of binding sites by using ChIP-Seq experiments and the value of reference samples that should be used in such experiments.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

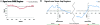
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

