Mechanism of ADP-ribosylation removal revealed by the structure and ligand complexes of the dimanganese mono-ADP-ribosylhydrolase DraG
- PMID: 19706507
- PMCID: PMC2732831
- DOI: 10.1073/pnas.0905906106
Mechanism of ADP-ribosylation removal revealed by the structure and ligand complexes of the dimanganese mono-ADP-ribosylhydrolase DraG
Abstract
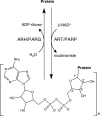
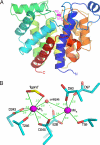
ADP-ribosylation is a ubiquitous regulatory posttranslational modification involved in numerous key processes such as DNA repair, transcription, cell differentiation, apoptosis, and the pathogenic mechanism of certain bacterial toxins. Despite the importance of this reversible process, very little is known about the structure and mechanism of the hydrolases that catalyze removal of the ADP-ribose moiety. In the phototrophic bacterium Rhodospirillum rubrum, dinitrogenase reductase-activating glycohydrolase (DraG), a dimanganese enzyme that reversibly associates with the cell membrane, is a key player in the regulation of nitrogenase activity. DraG has long served as a model protein for ADP-ribosylhydrolases. Here, we present the crystal structure of DraG in the holo and ADP-ribose bound forms. We also present the structure of a reaction intermediate analogue and propose a detailed catalytic mechanism for protein de-ADP-ribosylation involving ring opening of the substrate ribose. In addition, the particular manganese coordination in DraG suggests a rationale for the enzyme's preference for manganese over magnesium, although not requiring a redox active metal for the reaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
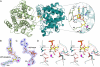

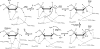
References
-
- Moss J, Zolkiewska A, Okazaki I. ADP-ribosylarginine hydrolases and ADP-ribosyltransferases. Partners in ADP-ribosylation cycles. Adv Exp Med Biol. 1997;419:25–33. - PubMed
-
- Koch-Nolte F, et al. Mammalian ADP-ribosyltransferases and ADP-ribosylhydolases. Front Biosci. 2008;13:6716–6729. - PubMed
-
- Deng Q, Barbieri JT. Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu Rev Microbiol. 2008;62:271–288. - PubMed
-
- Vanden Broeck D, Horvath C, De Wolf MJ. Vibrio cholerae: Cholera toxin. Int J Biochem Cell Biol. 2007;39:1771–1775. - PubMed
-
- Corda D, Di Girolamo M. Mono-ADP-ribosylation: A tool for modulating immune response and cell signaling. Sci STKE. 2002;2002:PE53. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

