Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis
- PMID: 19706706
- PMCID: PMC2772765
- DOI: 10.1128/JVI.00959-09
Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis
Abstract
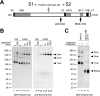
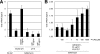
The coronaviruses mouse hepatitis virus type 2 (MHV-2) and severe acute respiratory syndrome coronavirus (SARS-CoV) utilize proteases to enter host cells. Upon receptor binding, the spike (S) proteins of both viruses are activated for membrane fusion by proteases, such as trypsin, present in the environment, facilitating virus entry from the cell surface. In contrast, in the absence of extracellular proteases, these viruses can enter cells via an endosomal pathway and utilize endosomal cathepsins for S protein activation. We demonstrate that the MHV-2 S protein uses multistep conformational changes for membrane fusion. After interaction with a soluble form of the MHV receptor (CEACAM1a), the metastable form of S protein is converted to a stable trimer, as revealed by mildly denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Liposome-binding assays indicate that the receptor-bound virions are associated with the target membrane through hydrophobic interactions. The exposure of receptor-bound S protein to trypsin or cathepsin L (CPL) induces the formation of six-helix bundles (6HB), the final conformation. This trypsin- or CPL-mediated conversion to 6HB can be blocked by a heptad repeat peptide known to block the formation of 6HB. Although trypsin treatment enabled receptor-bound MHV-2 to enter from the cell surface, CPL failed to do so. Interestingly, consecutive treatment with CPL and then chlorpromazine enabled a portion of the virus to enter from cell surface. These results suggest that trypsin suffices for the induction of membrane fusion of receptor-primed S protein, but an additional unidentified cellular factor is required to trigger membrane fusion by CPL.
Figures




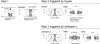
References
-
- Barnard, R. J. O., D. Elleder, and J. A. T. Young. 2006. Avian sarcoma and leukosis virus-receptor interactions: from classical genetics to novel insights into virus-cell membrane fusion. Virology 344:25-29. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

