Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice
- PMID: 19706712
- PMCID: PMC2753106
- DOI: 10.1128/JVI.00514-09
Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice
Abstract
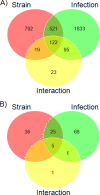
Despite the prevalence of H5N1 influenza viruses in global avian populations, comparatively few cases have been diagnosed in humans. Although viral factors almost certainly play a role in limiting human infection and disease, host genetics most likely contribute substantially. To model host factors in the context of influenza virus infection, we determined the lethal dose of a highly pathogenic H5N1 virus (A/Hong Kong/213/03) in C57BL/6J and DBA/2J mice and identified genetic elements associated with survival after infection. The lethal dose in these hosts varied by 4 logs and was associated with differences in replication kinetics and increased production of proinflammatory cytokines CCL2 and tumor necrosis factor alpha in susceptible DBA/2J mice. Gene mapping with recombinant inbred BXD strains revealed five loci or Qivr (quantitative trait loci for influenza virus resistance) located on chromosomes 2, 7, 11, 15, and 17 associated with resistance to H5N1 virus. In conjunction with gene expression profiling, we identified a number of candidate susceptibility genes. One of the validated genes, the hemolytic complement gene, affected virus titer 7 days after infection. We conclude that H5N1 influenza virus-induced pathology is affected by a complex and multigenic host component.
Figures
References
-
- Albright, F. S., P. Orlando, A. T. Pavia, G. G. Jackson, and L. A. C. Albright. 2008. Evidence for a heritable predisposition to death due to influenza. J. Infect. Dis. 197:18-24. - PubMed
-
- Alcais, A., M. Mira, J. L. Casanova, E. Schurr, and L. Abel. 2005. Genetic dissection of immunity in leprosy. Curr. Opin. Immunol. 17:44-48. - PubMed
-
- Aldridge, J. R., Jr., C. E. Moseley, D. A. Boltz, N. J. Negovetich, C. Reynolds, J. Franks, S. A. Brown, P. C. Doherty, R. G. Webster, and P. G. Thomas. 2009. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc. Natl. Acad. Sci. USA 106:5306-5311. - PMC - PubMed
-
- Balachandran, S., P. C. Roberts, L. E. Brown, H. Truong, A. K. Pattnaik, D. R. Archer, and G. N. Barber. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129-141. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

