Transcript profiling provides evidence of functional divergence and expression networks among ribosomal protein gene paralogs in Brassica napus
- PMID: 19706795
- PMCID: PMC2751962
- DOI: 10.1105/tpc.109.068411
Transcript profiling provides evidence of functional divergence and expression networks among ribosomal protein gene paralogs in Brassica napus
Abstract
The plant ribosome is composed of 80 distinct ribosomal (r)-proteins. In Arabidopsis thaliana, each r-protein is encoded by two or more highly similar paralogous genes, although only one copy of each r-protein is incorporated into the ribosome. Brassica napus is especially suited to the comparative study of r-protein gene paralogs due to its documented history of genome duplication as well as the recent availability of large EST data sets. We have identified 996 putative r-protein genes spanning 79 distinct r-proteins in B. napus using EST data from 16 tissue collections. A total of 23,408 tissue-specific r-protein ESTs are associated with this gene set. Comparative analysis of the transcript levels for these unigenes reveals that a large fraction of r-protein genes are differentially expressed and that the number of paralogs expressed for each r-protein varies extensively with tissue type in B. napus. In addition, in many cases the paralogous genes for a specific r-protein are not transcribed in concert and have highly contrasting expression patterns among tissues. Thus, each tissue examined has a novel r-protein transcript population. Furthermore, hierarchical clustering reveals that particular paralogs for nonhomologous r-protein genes cluster together, suggesting that r-protein paralog combinations are associated with specific tissues in B. napus and, thus, may contribute to tissue differentiation and/or specialization. Altogether, the data suggest that duplicated r-protein genes undergo functional divergence into highly specialized paralogs and coexpression networks and that, similar to recent reports for yeast, these are likely actively involved in differentiation, development, and/or tissue-specific processes.
Figures
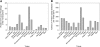
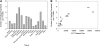

References
-
- Akashi, H. (2001). Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 11 660–666. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. - PubMed
-
- Aury, J.M., et al. (2006). Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444 171–178. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

