A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy
- PMID: 19706817
- PMCID: PMC7289055
- DOI: 10.1158/1078-0432.CCR-09-0113
A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy
Abstract
Purpose: Hans and coworkers previously developed an immunohistochemical algorithm with approximately 80% concordance with the gene expression profiling (GEP) classification of diffuse large B-cell lymphoma (DLBCL) into the germinal center B-cell-like (GCB) and activated B-cell-like (ABC) subtypes. Since then, new antibodies specific to germinal center B-cells have been developed, which might improve the performance of an immunostain algorithm.
Experimental design: We studied 84 cases of cyclophosphamide-doxorubicin-vincristine-prednisone (CHOP)-treated DLBCL (47 GCB, 37 ABC) with GCET1, CD10, BCL6, MUM1, FOXP1, BCL2, MTA3, and cyclin D2 immunostains, and compared different combinations of the immunostaining results with the GEP classification. A perturbation analysis was also applied to eliminate the possible effects of interobserver or intraobserver variations. A separate set of 63 DLBCL cases treated with rituximab plus CHOP (37 GCB, 26 ABC) was used to validate the new algorithm.
Results: A new algorithm using GCET1, CD10, BCL6, MUM1, and FOXP1 was derived that closely approximated the GEP classification with 93% concordance. Perturbation analysis indicated that the algorithm was robust within the range of observer variance. The new algorithm predicted 3-year overall survival of the validation set [GCB (87%) versus ABC (44%); P < 0.001], simulating the predictive power of the GEP classification. For a group of seven primary mediastinal large B-cell lymphoma, the new algorithm is a better prognostic classifier (all "GCB") than the Hans' algorithm (two GCB, five non-GCB).
Conclusion: Our new algorithm is significantly more accurate than the Hans' algorithm and will facilitate risk stratification of DLBCL patients and future DLBCL research using archival materials.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures
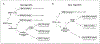

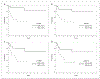

Comment in
-
Complexity made simple in diffuse large B-cell lymphoma.Clin Cancer Res. 2009 Sep 1;15(17):5291-3. doi: 10.1158/1078-0432.CCR-09-1444. Epub 2009 Aug 25. Clin Cancer Res. 2009. PMID: 19706818 Free PMC article.
-
Comparison of Choi and Hans' algorithms by immunohistochemistry and quantitative reverse transcriptase-PCR - letter.Clin Cancer Res. 2010 Jul 15;16(14):3805-6. doi: 10.1158/1078-0432.CCR-09-3216. Epub 2010 Jul 13. Clin Cancer Res. 2010. PMID: 20628029 No abstract available.
References
-
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403: 503–11. - PubMed
-
- Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 2002;346:1937–47. - PubMed
-
- A predictive model for aggressive non-Hodgkin's lymphoma. The International Non-Hodgkin's Lymphoma Prognostic Factors Project. N Engl J Med 1993;329:987–94. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

