European population genetic substructure: further definition of ancestry informative markers for distinguishing among diverse European ethnic groups
- PMID: 19707526
- PMCID: PMC2730349
- DOI: 10.2119/molmed.2009.00094
European population genetic substructure: further definition of ancestry informative markers for distinguishing among diverse European ethnic groups
Abstract
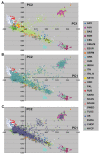
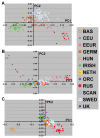
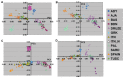
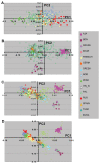
The definition of European population genetic substructure and its application to understanding complex phenotypes is becoming increasingly important. In the current study using over 4,000 subjects genotyped for 300,000 single-nucleotide polymorphisms (SNPs), we provide further insight into relationships among European population groups and identify sets of SNP ancestry informative markers (AIMs) for application in genetic studies. In general, the graphical description of these principal components analyses (PCA) of diverse European subjects showed a strong correspondence to the geographical relationships of specific countries or regions of origin. Clearer separation of different ethnic and regional populations was observed when northern and southern European groups were considered separately and the PCA results were influenced by the inclusion or exclusion of different self-identified population groups including Ashkenazi Jewish, Sardinian, and Orcadian ethnic groups. SNP AIM sets were identified that could distinguish the regional and ethnic population groups. Moreover, the studies demonstrated that most allele frequency differences between different European groups could be controlled effectively in analyses using these AIM sets. The European substructure AIMs should be widely applicable to ongoing studies to confirm and delineate specific disease susceptibility candidate regions without the necessity of performing additional genome-wide SNP studies in additional subject sets.
Figures

References
-
- Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

