Molecular models of the open and closed states of the whole human CFTR protein
- PMID: 19707853
- PMCID: PMC11115851
- DOI: 10.1007/s00018-009-0133-0
Molecular models of the open and closed states of the whole human CFTR protein
Abstract
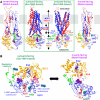
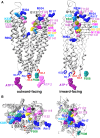
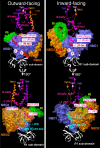
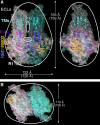
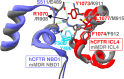
Cystic fibrosis transmembrane conductance regulator (CFTR), involved in cystic fibrosis (CF), is a chloride channel belonging to the ATP-binding cassette (ABC) superfamily. Using the experimental structure of Sav1866 as template, we previously modeled the human CFTR structure, including membrane-spanning domains (MSD) and nucleotide-binding domains (NBD), in an outward-facing conformation (open channel state). Here, we constructed a model of the CFTR inward-facing conformation (closed channel) on the basis of the recent corrected structures of MsbA and compared the structural features of those two states of the channel. Interestingly, the MSD:NBD coupling interfaces including F508 (DeltaF508 being the most common CF mutation) are mainly left unchanged. This prediction, completed by the modeling of the regulatory R domain, is supported by experimental data and provides a molecular basis for a better understanding of the functioning of CFTR, especially of the structural features that make CFTR the unique channel among the ABC transporters.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

