Identification of RNA helicase A as a new host factor in the replication cycle of foot-and-mouth disease virus
- PMID: 19710149
- PMCID: PMC2772801
- DOI: 10.1128/JVI.02677-08
Identification of RNA helicase A as a new host factor in the replication cycle of foot-and-mouth disease virus
Abstract
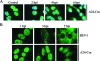
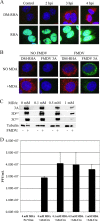
Foot-and-mouth disease virus (FMDV), as with other RNA viruses, recruits various host cell factors to assist in the translation and replication of the virus genome. In this study, we investigated the role of RNA helicase A (RHA) in the life cycle of FMDV. Immunofluorescent microscopy (IFM) showed a change in the subcellular distribution of RHA from the nucleus to the cytoplasm in FMDV-infected cells as infection progressed. Unlike nuclear RHA, the RHA detected in the cytoplasm reacted with an antibody that recognizes only the nonmethylated form of RHA. In contrast to alterations in the subcellular distribution of nuclear factors observed during infection with the related cardioviruses, cytoplasmic accumulation of RHA did not require the activity of the FMDV leader protein. Using IFM, we have found cytoplasmic RHA in proximity to the viral 2C and 3A proteins, which promotes the assembly of the replication complexes, as well as cellular poly(A) binding protein (PABP). Coimmunoprecipitation assays confirmed that these proteins are complexed with RHA. We have also identified a novel interaction between RHA and the S fragment in the FMDV 5' nontranslated region. Moreover, a reduction in the expression of RHA, using RHA-specific small interfering RNA constructs, inhibited FMDV replication. These results indicate that RHA plays an essential role in the replication of FMDV and potentially other picornaviruses through ribonucleoprotein complex formation at the 5' end of the genome and by interactions with 2C, 3A, and PABP.
Figures

Similar articles
-
Heterogeneous Nuclear Ribonucleoprotein L Negatively Regulates Foot-and-Mouth Disease Virus Replication through Inhibition of Viral RNA Synthesis by Interacting with the Internal Ribosome Entry Site in the 5' Untranslated Region.J Virol. 2020 May 4;94(10):e00282-20. doi: 10.1128/JVI.00282-20. Print 2020 May 4. J Virol. 2020. PMID: 32161169 Free PMC article.
-
The DDX23 Negatively Regulates Translation and Replication of Foot-and-Mouth Disease Virus and Is Degraded by 3C Proteinase.Viruses. 2020 Nov 25;12(12):1348. doi: 10.3390/v12121348. Viruses. 2020. PMID: 33255534 Free PMC article.
-
Heat Shock Protein 60 Is Involved in Viral Replication Complex Formation and Facilitates Foot and Mouth Virus Replication by Stabilizing Viral Nonstructural Proteins 3A and 2C.mBio. 2022 Oct 26;13(5):e0143422. doi: 10.1128/mbio.01434-22. Epub 2022 Sep 15. mBio. 2022. PMID: 36106732 Free PMC article.
-
The leader proteinase of foot-and-mouth disease virus: Efficiency through exosites.Virology. 2025 Mar;604:110420. doi: 10.1016/j.virol.2025.110420. Epub 2025 Jan 24. Virology. 2025. PMID: 39884163 Review.
-
Translation and replication of FMDV RNA.Curr Top Microbiol Immunol. 2005;288:43-70. doi: 10.1007/3-540-27109-0_3. Curr Top Microbiol Immunol. 2005. PMID: 15648174 Review.
Cited by
-
RNA Helicase A/DHX9 Forms Unique Cytoplasmic Antiviral Granules That Restrict Oncolytic Myxoma Virus Replication in Human Cancer Cells.J Virol. 2021 Jun 24;95(14):e0015121. doi: 10.1128/JVI.00151-21. Epub 2021 Jun 24. J Virol. 2021. PMID: 33952639 Free PMC article.
-
Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses.Virology. 2015 May;479-480:457-74. doi: 10.1016/j.virol.2015.03.001. Epub 2015 Mar 26. Virology. 2015. PMID: 25818028 Free PMC article. Review.
-
The pseudogene GBP1P1 suppresses influenza A virus replication by acting as a protein decoy for DHX9.J Virol. 2024 Jul 23;98(7):e0073824. doi: 10.1128/jvi.00738-24. Epub 2024 Jun 28. J Virol. 2024. PMID: 38940585 Free PMC article.
-
The expanding functions of cellular helicases: the tombusvirus RNA replication enhancer co-opts the plant eIF4AIII-like AtRH2 and the DDX5-like AtRH5 DEAD-box RNA helicases to promote viral asymmetric RNA replication.PLoS Pathog. 2014 Apr 17;10(4):e1004051. doi: 10.1371/journal.ppat.1004051. eCollection 2014 Apr. PLoS Pathog. 2014. PMID: 24743583 Free PMC article.
-
Dynamically probing ATP-dependent RNA helicase A-assisted RNA structure conversion using single molecule fluorescence resonance energy transfer.Protein Sci. 2021 Jun;30(6):1157-1168. doi: 10.1002/pro.4081. Protein Sci. 2021. PMID: 33837988 Free PMC article.
References
-
- Alvarez, D. E., C. V. Filomatori, and A. V. Gamarnik. 2008. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology 375:223-235. - PubMed
-
- Alvarez, D. E., M. F. Lodeiro, C. V. Filomatori, S. Fucito, J. A. Mondotte, and A. V. Gamarnik. 2006. Structural and functional analysis of dengue virus RNA. Novartis Found. Symp. 277:120-132. - PubMed
-
- Andino, R., N. Boddeker, D. Silvera, and A. V. Gamarnik. 1999. Intracellular determinants of picornavirus replication. Trends Microbiol. 7:76-82. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

