Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbalpha
- PMID: 19710360
- PMCID: PMC2751986
- DOI: 10.1101/gad.1825809
Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbalpha
Abstract
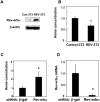
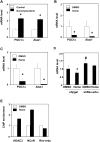
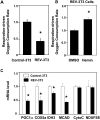
Intracellular heme levels must be tightly regulated to maintain proper mitochondrial respiration while minimizing toxicity, but the homeostatic mechanisms are not well understood. Here we report a novel negative feedback mechanism whereby the nuclear heme receptor Rev-erbalpha tightly controls the level of its own ligand. Heme binding to Rev-erbalpha recruits the NCoR/histone deacetylase 3 (HDAC3) corepressor complex to repress the transcription of the coactivator PGC-1alpha, a potent inducer of heme synthesis. Depletion of Rev-erbalpha derepresses PGC-1alpha, resulting in increased heme levels. Conversely, increased Rev-erbalpha reduces intracellular heme, and impairs mitochondrial respiration in a heme-dependent manner. Consistent with this bioenergetic impairment, overexpression of Rev-erbalpha dramatically inhibits cell growth due to a cell cycle arrest. Thus, Rev-erbalpha modulates the synthesis of its own ligand in a negative feedback pathway that maintains heme levels and regulates cellular energy metabolism.
Figures




Similar articles
-
PGC-1alpha negatively regulates hepatic FGF21 expression by modulating the heme/Rev-Erb(alpha) axis.Proc Natl Acad Sci U S A. 2009 Dec 29;106(52):22510-5. doi: 10.1073/pnas.0912533106. Epub 2009 Dec 14. Proc Natl Acad Sci U S A. 2009. PMID: 20018698 Free PMC article.
-
Relationship between circadian oscillations of Rev-erbalpha expression and intracellular levels of its ligand, heme.Biochem Biophys Res Commun. 2008 Apr 18;368(4):955-8. doi: 10.1016/j.bbrc.2008.02.031. Epub 2008 Feb 15. Biochem Biophys Res Commun. 2008. PMID: 18280802 Free PMC article.
-
The nuclear receptor Rev-erbalpha is a liver X receptor (LXR) target gene driving a negative feedback loop on select LXR-induced pathways in human macrophages.Mol Endocrinol. 2008 Aug;22(8):1797-811. doi: 10.1210/me.2007-0439. Epub 2008 May 29. Mol Endocrinol. 2008. PMID: 18511497 Free PMC article.
-
The orphan nuclear receptor Rev-erbalpha: a transcriptional link between circadian rhythmicity and cardiometabolic disease.Curr Opin Lipidol. 2007 Apr;18(2):141-6. doi: 10.1097/MOL.0b013e3280464ef6. Curr Opin Lipidol. 2007. PMID: 17353661 Review.
-
Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock.Mol Endocrinol. 2008 Jul;22(7):1509-20. doi: 10.1210/me.2007-0519. Epub 2008 Jan 24. Mol Endocrinol. 2008. PMID: 18218725 Free PMC article. Review.
Cited by
-
DBC1 (Deleted in Breast Cancer 1) modulates the stability and function of the nuclear receptor Rev-erbα.Biochem J. 2013 May 1;451(3):453-61. doi: 10.1042/BJ20121085. Biochem J. 2013. PMID: 23398316 Free PMC article.
-
Clocks, metabolism, and the epigenome.Mol Cell. 2012 Jul 27;47(2):158-67. doi: 10.1016/j.molcel.2012.06.026. Mol Cell. 2012. PMID: 22841001 Free PMC article. Review.
-
Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy.Nat Med. 2013 Aug;19(8):1039-46. doi: 10.1038/nm.3213. Epub 2013 Jul 14. Nat Med. 2013. PMID: 23852339 Free PMC article.
-
The melatonin agonist ramelteon induces duration-dependent clock gene expression through cAMP signaling in pancreatic INS-1 β-cells.PLoS One. 2014 Jul 11;9(7):e102073. doi: 10.1371/journal.pone.0102073. eCollection 2014. PLoS One. 2014. PMID: 25013953 Free PMC article.
-
Drosophila Evi5 is a critical regulator of intracellular iron transport via transferrin and ferritin interactions.Nat Commun. 2024 May 14;15(1):4045. doi: 10.1038/s41467-024-48165-9. Nat Commun. 2024. PMID: 38744835 Free PMC article.
References
-
- Atamna H. Heme, iron, and the mitochondrial decay of ageing. Ageing Res Rev. 2004;3:303–318. - PubMed
-
- Carroll JS, Brown M. Estrogen receptor target gene: An evolving concept. Mol Endocrinol. 2006;20:1707–1714. - PubMed
-
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. - PubMed
-
- Chiamolera MI, Wondisford FE. Minireview: Thyrotropin-releasing hormone and the thyroid hormone feedback mechanism. Endocrinology. 2009;150:1091–1096. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
