microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells
- PMID: 19710908
- PMCID: PMC2728509
- DOI: 10.1371/journal.pone.0006783
microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells
Abstract
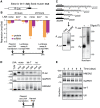
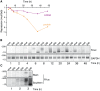
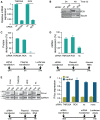
Animal microRNAs (miRNAs) typically regulate gene expression by binding to partially complementary target sites in the 3' untranslated region (UTR) of messenger RNA (mRNA) reducing its translation and stability. They also commonly induce shortening of the mRNA 3' poly(A) tail, which contributes to their mRNA decay promoting function. The relationship between miRNA-mediated deadenylation and translational repression has been less clear. Using transfection of reporter constructs carrying three imperfectly matching let-7 target sites in the 3' UTR into mammalian cells we observe rapid target mRNA deadenylation that precedes measureable translational repression by endogenous let-7 miRNA. Depleting cells of the argonaute co-factors RCK or TNRC6A can impair let-7-mediated repression despite ongoing mRNA deadenylation, indicating that deadenylation alone is not sufficient to effect full repression. Nevertheless, the magnitude of translational repression by let-7 is diminished when the target reporter lacks a poly(A) tail. Employing an antisense strategy to block deadenylation of target mRNA with poly(A) tail also partially impairs translational repression. On the one hand, these experiments confirm that tail removal by deadenylation is not strictly required for translational repression. On the other hand they show directly that deadenylation can augment miRNA-mediated translational repression in mammalian cells beyond stimulating mRNA decay. Taken together with published work, these results suggest a dual role of deadenylation in miRNA function: it contributes to translational repression as well as mRNA decay and is thus critically involved in establishing the quantitatively appropriate physiological response to miRNAs.
Conflict of interest statement
Figures


References
-
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. - PubMed
-
- Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8:215–239. - PubMed
-
- Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–623. - PubMed
-
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

