Arborization pattern of engrailed-positive neural lineages reveal neuromere boundaries in the Drosophila brain neuropil
- PMID: 19711412
- PMCID: PMC2879895
- DOI: 10.1002/cne.22112
Arborization pattern of engrailed-positive neural lineages reveal neuromere boundaries in the Drosophila brain neuropil
Abstract
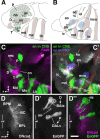
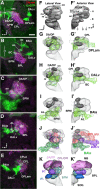
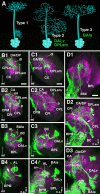
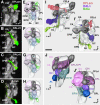
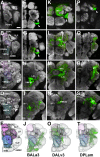
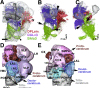
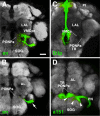
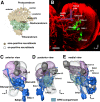
The Drosophila brain is a highly complex structure composed of thousands of neurons that are interconnected in numerous exquisitely organized neuropil structures such as the mushroom bodies, central complex, antennal lobes, and other specialized neuropils. While the neurons of the insect brain are known to derive in a lineage-specific fashion from a stereotyped set of segmentally organized neuroblasts, the developmental origin and neuromeric organization of the neuropil formed by these neurons is still unclear. In this study we used genetic labeling techniques to characterize the neuropil innervation pattern of engrailed-expressing brain lineages of known neuromeric origin. We show that the neurons of these lineages project to and form most arborizations, in particular all of their proximal branches, in the same brain neuropil compartments in embryonic, larval and adult stages. Moreover, we show that engrailed-positive neurons of differing neuromeric origin respect boundaries between neuromere-specific compartments in the brain. This is confirmed by an analysis of the arborization pattern of empty spiracles-expressing lineages. These findings indicate that arborizations of lineages deriving from different brain neuromeres innervate a nonoverlapping set of neuropil compartments. This supports a model for neuromere-specific brain neuropil, in which a given lineage forms its proximal arborizations predominantly in the compartments that correspond to its neuromere of origin.
Figures
Similar articles
-
The insect central complex as model for heterochronic brain development-background, concepts, and tools.Dev Genes Evol. 2016 Jun;226(3):209-19. doi: 10.1007/s00427-016-0542-7. Epub 2016 Apr 7. Dev Genes Evol. 2016. PMID: 27056385 Free PMC article. Review.
-
Structure and development of the subesophageal zone of the Drosophila brain. II. Sensory compartments.J Comp Neurol. 2018 Jan 1;526(1):33-58. doi: 10.1002/cne.24316. Epub 2017 Sep 28. J Comp Neurol. 2018. PMID: 28875566 Free PMC article.
-
Structure and development of the subesophageal zone of the Drosophila brain. I. Segmental architecture, compartmentalization, and lineage anatomy.J Comp Neurol. 2018 Jan 1;526(1):6-32. doi: 10.1002/cne.24287. Epub 2017 Aug 10. J Comp Neurol. 2018. PMID: 28730682 Free PMC article.
-
Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage.J Neurosci. 2006 May 17;26(20):5534-53. doi: 10.1523/JNEUROSCI.4708-05.2006. J Neurosci. 2006. PMID: 16707805 Free PMC article.
-
Lineages to circuits: the developmental and evolutionary architecture of information channels into the central complex.J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2023 Jul;209(4):679-720. doi: 10.1007/s00359-023-01616-y. Epub 2023 Mar 17. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2023. PMID: 36932234 Free PMC article. Review.
Cited by
-
The insect central complex as model for heterochronic brain development-background, concepts, and tools.Dev Genes Evol. 2016 Jun;226(3):209-19. doi: 10.1007/s00427-016-0542-7. Epub 2016 Apr 7. Dev Genes Evol. 2016. PMID: 27056385 Free PMC article. Review.
-
A small cohort of FRU(M) and Engrailed-expressing neurons mediate successful copulation in Drosophila melanogaster.BMC Neurosci. 2013 May 21;14:57. doi: 10.1186/1471-2202-14-57. BMC Neurosci. 2013. PMID: 23688386 Free PMC article.
-
Origin and development of neuropil glia of the Drosophila larval and adult brain: Two distinct glial populations derived from separate progenitors.Dev Biol. 2015 Aug 15;404(2):2-20. doi: 10.1016/j.ydbio.2015.03.004. Epub 2015 Mar 14. Dev Biol. 2015. PMID: 25779704 Free PMC article.
-
Structure and development of the subesophageal zone of the Drosophila brain. II. Sensory compartments.J Comp Neurol. 2018 Jan 1;526(1):33-58. doi: 10.1002/cne.24316. Epub 2017 Sep 28. J Comp Neurol. 2018. PMID: 28875566 Free PMC article.
-
Patterns of growth and tract formation during the early development of secondary lineages in the Drosophila larval brain.Dev Neurobiol. 2016 Apr;76(4):434-51. doi: 10.1002/dneu.22325. Epub 2015 Jul 28. Dev Neurobiol. 2016. PMID: 26178322 Free PMC article.
References
-
- Baba Y, Comer CM. Antennal motor system of the cockroach, Periplaneta americana. Cell Tissue Res. 2008;331:751–62. - PubMed
-
- Beaster-Jones L, Kaltenbach SL, Koop D, Yuan S, Chastain R, Holland LZ. Expression of somite segmentation genes in amphioxus: a clock without a wavefront? Dev Genes Evol. 2008;218:599–611. - PubMed
-
- Bello BC, H. F, Gould AP. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–19. - PubMed
-
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. - PubMed
-
- Boyan G, Williams L. A single cell analysis of engrailed expression in the early embryonic brain of the grasshopper Schistocerca gregaria: ontogeny and identity of the secondary headspot cells. Arthropod Struct Dev. 2002;30:207–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

