Transcriptional profiling reveals developmental relationship and distinct biological functions of CD16+ and CD16- monocyte subsets
- PMID: 19712453
- PMCID: PMC2741492
- DOI: 10.1186/1471-2164-10-403
Transcriptional profiling reveals developmental relationship and distinct biological functions of CD16+ and CD16- monocyte subsets
Abstract
Background: Human peripheral blood monocytes (Mo) consist of subsets distinguished by expression of CD16 (FCgammaRIII) and chemokine receptors. Classical CD16- Mo express CCR2 and migrate in response to CCL2, while a minor CD16+ Mo subset expresses CD16 and CX3CR1 and migrates into tissues expressing CX3CL1. CD16+ Mo produce pro-inflammatory cytokines and are expanded in certain inflammatory conditions including sepsis and HIV infection.
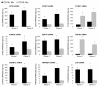
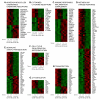
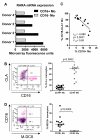
Results: To gain insight into the developmental relationship and functions of CD16+ and CD16- Mo, we examined transcriptional profiles of these Mo subsets in peripheral blood from healthy individuals. Of 16,328 expressed genes, 2,759 genes were differentially expressed and 228 and 250 were >2-fold upregulated and downregulated, respectively, in CD16+ compared to CD16- Mo. CD16+ Mo were distinguished by upregulation of transcripts for dendritic cell (DC) (SIGLEC10, CD43, RARA) and macrophage (MPhi) (CSF1R/CD115, MafB, CD97, C3aR) markers together with transcripts relevant for DC-T cell interaction (CXCL16, ICAM-2, LFA-1), cell activation (LTB, TNFRSF8, LST1, IFITM1-3, HMOX1, SOD-1, WARS, MGLL), and negative regulation of the cell cycle (CDKN1C, MTSS1), whereas CD16- Mo were distinguished by upregulation of transcripts for myeloid (CD14, MNDA, TREM1, CD1d, C1qR/CD93) and granulocyte markers (FPR1, GCSFR/CD114, S100A8-9/12). Differential expression of CSF1R, CSF3R, C1QR1, C3AR1, CD1d, CD43, CXCL16, and CX3CR1 was confirmed by flow cytometry. Furthermore, increased expression of RARA and KLF2 transcripts in CD16+ Mo coincided with absence of cell surface cutaneous lymphocyte associated antigen (CLA) expression, indicating potential imprinting for non-skin homing.
Conclusion: These results suggest that CD16+ and CD16- Mo originate from a common myeloid precursor, with CD16+ Mo having a more MPhi - and DC-like transcription program suggesting a more advanced stage of differentiation. Distinct transcriptional programs, together with their recruitment into tissues via different mechanisms, also suggest that CD16+ and CD16- Mo give rise to functionally distinct DC and MPhi in vivo.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

