Regulation of immature dendritic cell migration by RhoA guanine nucleotide exchange factor Arhgef5
- PMID: 19713215
- PMCID: PMC2781403
- DOI: 10.1074/jbc.M109.047282
Regulation of immature dendritic cell migration by RhoA guanine nucleotide exchange factor Arhgef5
Abstract
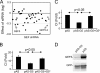
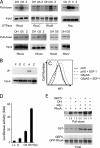
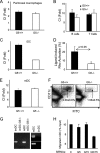
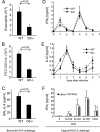
There are a large number of Rho guanine nucleotide exchange factors, most of which have no known functions. Here, we carried out a short hairpin RNA-based functional screen of Rho-GEFs for their roles in leukocyte chemotaxis and identified Arhgef5 as an important factor in chemotaxis of a macrophage phage-like RAW264.7 cell line. Arhgef5 can strongly activate RhoA and RhoB and weakly RhoC and RhoG, but not Rac1, RhoQ, RhoD, or RhoV, in transfected human embryonic kidney 293 cells. In addition, Gbetagamma interacts with Arhgef5 and can stimulate Arhgef5-mediated activation of RhoA in an in vitro assay. In vivo roles of Arhgef5 were investigated using an Arhgef-5-null mouse line. Arhgef5 deficiency did not affect chemotaxis of mouse macrophages, T and B lymphocytes, and bone marrow-derived mature dendritic cells (DC), but it abrogated MIP1alpha-induced chemotaxis of immature DCs and impaired migration of DCs from the skin to lymph node. In addition, Arhgef5 deficiency attenuated allergic airway inflammation. Therefore, this study provides new insights into signaling mechanisms for DC migration regulation.
Figures

Similar articles
-
Odontogenic Ameloblast-associated Protein (ODAM) Mediates Junctional Epithelium Attachment to Teeth via Integrin-ODAM-Rho Guanine Nucleotide Exchange Factor 5 (ARHGEF5)-RhoA Signaling.J Biol Chem. 2015 Jun 5;290(23):14740-53. doi: 10.1074/jbc.M115.648022. Epub 2015 Apr 24. J Biol Chem. 2015. PMID: 25911094 Free PMC article.
-
XPLN, a guanine nucleotide exchange factor for RhoA and RhoB, but not RhoC.J Biol Chem. 2002 Nov 8;277(45):42964-72. doi: 10.1074/jbc.M207401200. Epub 2002 Sep 6. J Biol Chem. 2002. PMID: 12221096
-
Expression and molecular characterization of alternative transcripts of the ARHGEF5/TIM oncogene specific for human breast cancer.Hum Mol Genet. 2004 Feb 1;13(3):323-34. doi: 10.1093/hmg/ddh024. Epub 2003 Dec 8. Hum Mol Genet. 2004. PMID: 14662653
-
The guanine nucleotide exchange factor Tiam1: a Janus-faced molecule in cellular signaling.Cell Signal. 2014 Mar;26(3):483-91. doi: 10.1016/j.cellsig.2013.11.034. Epub 2013 Dec 2. Cell Signal. 2014. PMID: 24308970 Review.
-
A current overview of RhoA, RhoB, and RhoC functions in vascular biology and pathology.Biochem Pharmacol. 2022 Dec;206:115321. doi: 10.1016/j.bcp.2022.115321. Epub 2022 Oct 25. Biochem Pharmacol. 2022. PMID: 36306821 Review.
Cited by
-
Odontogenic Ameloblast-associated Protein (ODAM) Mediates Junctional Epithelium Attachment to Teeth via Integrin-ODAM-Rho Guanine Nucleotide Exchange Factor 5 (ARHGEF5)-RhoA Signaling.J Biol Chem. 2015 Jun 5;290(23):14740-53. doi: 10.1074/jbc.M115.648022. Epub 2015 Apr 24. J Biol Chem. 2015. PMID: 25911094 Free PMC article.
-
RhoA as a Key Regulator of Innate and Adaptive Immunity.Cells. 2019 Jul 17;8(7):733. doi: 10.3390/cells8070733. Cells. 2019. PMID: 31319592 Free PMC article. Review.
-
A microfluidic-based genetic screen to identify microbial virulence factors that inhibit dendritic cell migration.Integr Biol (Camb). 2014 Apr;6(4):438-49. doi: 10.1039/c3ib40177d. Epub 2014 Mar 6. Integr Biol (Camb). 2014. PMID: 24599496 Free PMC article.
-
MicroRNA Regulation of the Small Rho GTPase Regulators-Complexities and Opportunities in Targeting Cancer Metastasis.Cancers (Basel). 2020 Apr 28;12(5):1092. doi: 10.3390/cancers12051092. Cancers (Basel). 2020. PMID: 32353968 Free PMC article. Review.
-
Distinct RhoGEFs Activate Apical and Junctional Contractility under Control of G Proteins during Epithelial Morphogenesis.Curr Biol. 2019 Oct 21;29(20):3370-3385.e7. doi: 10.1016/j.cub.2019.08.017. Epub 2019 Sep 12. Curr Biol. 2019. PMID: 31522942 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

