Chromatin poises miRNA- and protein-coding genes for expression
- PMID: 19713549
- PMCID: PMC2765269
- DOI: 10.1101/gr.090951.109
Chromatin poises miRNA- and protein-coding genes for expression
Erratum in
- Genome Res. 2009 Dec;19(12):2343
Abstract
Chromatin modifications have been implicated in the regulation of gene expression. While association of certain modifications with expressed or silent genes has been established, it remains unclear how changes in chromatin environment relate to changes in gene expression. In this article, we used ChIP-seq (chromatin immunoprecipitation with massively parallel sequencing) to analyze the genome-wide changes in chromatin modifications during activation of total human CD4(+) T cells by T-cell receptor (TCR) signaling. Surprisingly, we found that the chromatin modification patterns at many induced and silenced genes are relatively stable during the short-term activation of resting T cells. Active chromatin modifications were already in place for a majority of inducible protein-coding genes, even while the genes were silent in resting cells. Similarly, genes that were silenced upon T-cell activation retained positive chromatin modifications even after being silenced. To investigate if these observations are also valid for miRNA-coding genes, we systematically identified promoters for known miRNA genes using epigenetic marks and profiled their expression patterns using deep sequencing. We found that chromatin modifications can poise miRNA-coding genes as well. Our data suggest that miRNA- and protein-coding genes share similar mechanisms of regulation by chromatin modifications, which poise inducible genes for activation in response to environmental stimuli.
Figures

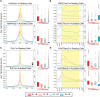


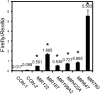

References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Bartel DP, Chen CZ. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. - PubMed
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. - PubMed
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
