Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil
- PMID: 19714218
- PMCID: PMC2726614
- DOI: 10.1371/journal.pgen.1000621
Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil
Erratum in
-
Correction: Penetration of the Stigma and Style Elicits a Novel Transcriptome in Pollen Tubes, Pointing to Genes Critical for Growth in a Pistil.PLoS Genet. 2016 Jul 19;12(7):e1006210. doi: 10.1371/journal.pgen.1006210. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27434655 Free PMC article.
Abstract
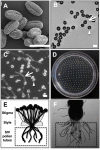
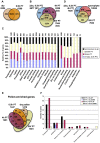
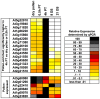
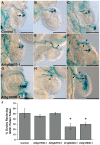
Pollen tubes extend through pistil tissues and are guided to ovules where they release sperm for fertilization. Although pollen tubes can germinate and elongate in a synthetic medium, their trajectory is random and their growth rates are slower compared to growth in pistil tissues. Furthermore, interaction with the pistil renders pollen tubes competent to respond to guidance cues secreted by specialized cells within the ovule. The molecular basis for this potentiation of the pollen tube by the pistil remains uncharacterized. Using microarray analysis in Arabidopsis, we show that pollen tubes that have grown through stigma and style tissues of a pistil have a distinct gene expression profile and express a substantially larger fraction of the Arabidopsis genome than pollen grains or pollen tubes grown in vitro. Genes involved in signal transduction, transcription, and pollen tube growth are overrepresented in the subset of the Arabidopsis genome that is enriched in pistil-interacted pollen tubes, suggesting the possibility of a regulatory network that orchestrates gene expression as pollen tubes migrate through the pistil. Reverse genetic analysis of genes induced during pollen tube growth identified seven that had not previously been implicated in pollen tube growth. Two genes are required for pollen tube navigation through the pistil, and five genes are required for optimal pollen tube elongation in vitro. Our studies form the foundation for functional genomic analysis of the interactions between the pollen tube and the pistil, which is an excellent system for elucidation of novel modes of cell-cell interaction.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Bishopp A, Mahonen AP, Helariutta Y. Signs of change: hormone receptors that regulate plant development. Development. 2006;133:1857–1869. - PubMed
-
- Borg M, Brownfield L, Twell D. Male gametophyte development: a molecular perspective. J Exp Bot. 2009;60:1465–1478. - PubMed
-
- von Besser K, Frank AC, Johnson MA, Preuss D. Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development. 2006;133:4761–4769. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

