Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle
- PMID: 19714224
- PMCID: PMC2727004
- DOI: 10.1371/journal.ppat.1000565
Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle
Abstract
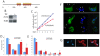
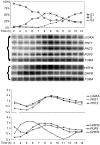
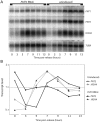
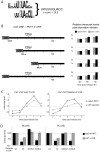
Many genes that are required at specific points in the cell cycle exhibit cell cycle-dependent expression. In the early-diverging model eukaryote and important human pathogen Trypanosoma brucei, regulation of gene expression in the cell cycle and other processes is almost entirely post-transcriptional. Here, we show that the T. brucei RNA-binding protein PUF9 stabilizes certain transcripts during S-phase. Target transcripts of PUF9--LIGKA, PNT1 and PNT2--were identified by affinity purification with TAP-tagged PUF9. RNAi against PUF9 caused an accumulation of cells in G2/M phase and unexpectedly destabilized the PUF9 target mRNAs, despite the fact that most known Puf-domain proteins promote degradation of their target mRNAs. The levels of the PUF9-regulated transcripts were cell cycle dependent, peaking in mid- to late- S-phase, and this effect was abolished when PUF9 was targeted by RNAi. The sequence UUGUACC was over-represented in the 3' UTRs of PUF9 targets; a point mutation in this motif abolished PUF9-dependent stabilization of a reporter transcript carrying the PNT1 3' UTR. LIGKA is involved in replication of the kinetoplast, and here we show that PNT1 is also kinetoplast-associated and its over-expression causes kinetoplast-related defects, while PNT2 is localized to the nucleus in G1 phase and redistributes to the mitotic spindle during mitosis. PUF9 targets may constitute a post-transcriptional regulon, encoding proteins involved in temporally coordinated replicative processes in early G2 phase.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Johnson LF, Fuhrman CL, Wiedemann LM. Regulation of dihydrofolate reductase gene expression in mouse fibroblasts during the transition from the resting to growing state. J Cell Physiol. 1978;97:397–306. - PubMed
-
- Stein G, Park W, Thrall C, Mans R, Stein J. Regulation of cell cycle stage-specific transcription of histone genes from chromatin by non-histone chromosomal proteins. Nature. 1975;257:764–767. - PubMed
-
- Noe V, Ciudad CJ, Chasin LA. Effect of differential polyadenylation and cell growth phase on dihydrofolate reductase mRNA stability. J Biol Chem. 1999;274:27807–27814. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

