Assessment of ligand binding residue predictions in CASP8
- PMID: 19714771
- PMCID: PMC3204792
- DOI: 10.1002/prot.22557
Assessment of ligand binding residue predictions in CASP8
Abstract
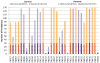
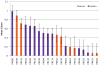
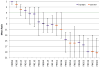
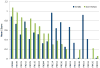
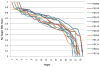
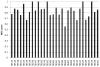
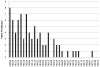
Here we detail the assessment process for the binding site prediction category of the eighth Critical Assessment of Protein Structure Prediction experiment (CASP8). Predictions were only evaluated for those targets that bound biologically relevant ligands and were assessed using the Matthews Correlation Coefficient. The results of the analysis clearly demonstrate that three predictors from two groups (Lee and Sternberg) stand out from the rest. A further two groups perform well over subsets of metal binding or nonmetal ligand binding targets. The best methods were able to make consistently reliable predictions based on model structures, though it was noticeable that the two targets that were not well predicted were also the hardest targets. The number of predictors that submitted new methods in this category was highly encouraging and suggests that current technology is at the level that experimental biochemists and structural biologists could benefit from what is clearly a growing field.
Copyright 2009 Wiley-Liss, Inc.
Figures

References
-
- Gherardini PF, Helmer-Citterich M. Structure-based function prediction: approaches and applications. Brief Funct Genomic Proteomic. 2008;7(4):291–302. - PubMed
-
- Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20(8):1322–1324. - PubMed
-
- Casari G, Sander C, Valencia A. A method to predict functional residues in proteins. Nat Struct Biol. 1995;2(2):171–178. - PubMed
-
- del Sol A, Pazos F, Valencia A. Automatic methods for predicting functionally important residues. J Mol Biol. 2003;326(4):1289–1302. - PubMed
-
- Fischer JD, Mayer CE, Soding J. Prediction of protein functional residues from sequence by probability density estimation. Bioinformatics. 2008;24(5):613–620. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

