TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres
- PMID: 19716786
- PMCID: PMC2749977
- DOI: 10.1016/j.molcel.2009.06.025
TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres
Abstract
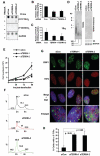
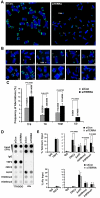
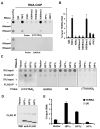
Telomere-repeat-encoding RNA (referred to as TERRA) has been identified as a potential component of yeast and mammalian telomeres. We show here that TERRA RNA interacts with several telomere-associated proteins, including telomere repeat factors 1 (TRF1) and 2 (TRF2), subunits of the origin recognition complex (ORC), heterochromatin protein 1 (HP1), histone H3 trimethyl K9 (H3 K9me3), and members of the DNA-damage-sensing pathway. siRNA depletion of TERRA caused an increase in telomere dysfunction-induced foci, aberrations in metaphase telomeres, and a loss of histone H3 K9me3 and ORC at telomere repeat DNA. Previous studies found that TRF2 amino-terminal GAR domain recruited ORC to telomeres. We now show that TERRA RNA can interact directly with the TRF2 GAR and ORC1 to form a stable ternary complex. We conclude that TERRA facilitates TRF2 interaction with ORC and plays a central role in telomere structural maintenance and heterochromatin formation.
Figures




References
-
- Atanasiu C, Lezina L, Lieberman PM. DNA affinity purification of Epstein-Barr virus OriP-binding proteins. Methods Mol Biol. 2005;292:267–276. - PubMed
-
- Azzalin CM, Reichenback P, Khoriauli L, Giulotto E, Lingner J. Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science. 2007;318:798–801. - PubMed
-
- Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–672. - PubMed
-
- Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–1138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

