Single-residue changes in the C-terminal disulfide-bonded loop of the Pseudomonas aeruginosa type IV pilin influence pilus assembly and twitching motility
- PMID: 19717595
- PMCID: PMC2795284
- DOI: 10.1128/JB.00943-09
Single-residue changes in the C-terminal disulfide-bonded loop of the Pseudomonas aeruginosa type IV pilin influence pilus assembly and twitching motility
Abstract
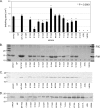
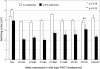
PilA, the major pilin subunit of Pseudomonas aeruginosa type IV pili (T4P), is a principal structural component. PilA has a conserved C-terminal disulfide-bonded loop (DSL) that has been implicated as the pilus adhesinotope. Structural studies have suggested that DSL is involved in intersubunit interactions within the pilus fiber. PilA mutants with single-residue substitutions, insertions, or deletions in the DSL were tested for pilin stability, pilus assembly, and T4P function. Mutation of either Cys residue of the DSL resulted in pilins that were unable to assemble into fibers. Ala replacements of the intervening residues had a range of effects on assembly or function, as measured by changes in surface pilus expression and twitching motility. Modification of the C-terminal P-X-X-C type II beta-turn motif, which is one of the few highly conserved features in pilins across various species, caused profound defects in assembly and twitching motility. Expression of pilins with suspected assembly defects in a pilA pilT double mutant unable to retract T4P allowed us to verify which subunits were physically unable to assemble. Use of two different PilA antibodies showed that the DSL may be an immunodominant epitope in intact pili compared with pilin monomers. Sequence diversity of the type IVa pilins likely reflects an evolutionary compromise between retention of function and antigenic variation. The consequences of DSL sequence changes should be evaluated in the intact protein since it is technically feasible to generate DSL-mimetic peptides with mutations that will not appear in the natural repertoire due to their deleterious effects on assembly.
Figures




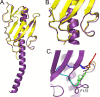
Similar articles
-
Structural and functional studies of the Pseudomonas aeruginosa minor pilin, PilE.J Biol Chem. 2015 Oct 30;290(44):26856-65. doi: 10.1074/jbc.M115.683334. Epub 2015 Sep 10. J Biol Chem. 2015. PMID: 26359492 Free PMC article.
-
Structural conservation and functional role of TfpY-like proteins in type IV pilus assembly.J Bacteriol. 2025 Feb 20;207(2):e0034324. doi: 10.1128/jb.00343-24. Epub 2025 Jan 16. J Bacteriol. 2025. PMID: 39817748 Free PMC article.
-
Structural characterization of novel Pseudomonas aeruginosa type IV pilins.J Mol Biol. 2010 Jan 22;395(3):491-503. doi: 10.1016/j.jmb.2009.10.070. Epub 2009 Nov 4. J Mol Biol. 2010. PMID: 19895819
-
The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa--a review.Gene. 1997 Jun 11;192(1):99-108. doi: 10.1016/s0378-1119(97)00116-9. Gene. 1997. PMID: 9224879 Review.
-
Type IV pili: dynamics, biophysics and functional consequences.Nat Rev Microbiol. 2019 Jul;17(7):429-440. doi: 10.1038/s41579-019-0195-4. Nat Rev Microbiol. 2019. PMID: 30988511 Review.
Cited by
-
Understanding a Core Pilin of the Type IVa Pili of Acidithiobacillus thiooxidans, PilV.J Microbiol Biotechnol. 2024 Mar 28;34(3):527-537. doi: 10.4014/jmb.2310.10033. Epub 2024 Jan 17. J Microbiol Biotechnol. 2024. PMID: 38346803 Free PMC article.
-
Nanoscale Pulling of Type IV Pili Reveals Their Flexibility and Adhesion to Surfaces over Extended Lengths of the Pili.Biophys J. 2015 Jun 16;108(12):2865-75. doi: 10.1016/j.bpj.2015.05.016. Biophys J. 2015. PMID: 26083926 Free PMC article.
-
Modeling and Simulating the Dynamics of Type IV Pili Extension of Pseudomonas aeruginosa.Biophys J. 2016 Nov 15;111(10):2263-2273. doi: 10.1016/j.bpj.2016.09.050. Biophys J. 2016. PMID: 27851948 Free PMC article.
-
Phage Resistance Evolution Induces the Sensitivity of Specific Antibiotics in Pseudomonas aeruginosa PAO1.Microbiol Spectr. 2022 Oct 26;10(5):e0135622. doi: 10.1128/spectrum.01356-22. Epub 2022 Aug 16. Microbiol Spectr. 2022. PMID: 35972274 Free PMC article.
-
Genomic and proteomic characterization of the large Myoviridae bacteriophage ϕTMA of the extreme thermophile Thermus thermophilus.Bacteriophage. 2011 May;1(3):152-164. doi: 10.4161/bact.1.3.16712. Epub 2011 May 1. Bacteriophage. 2011. PMID: 22164349 Free PMC article.
References
-
- Aas, F. E., H. C. Winther-Larsen, M. Wolfgang, S. Frye, C. Lovold, N. Roos, J. P. van Putten, and M. Koomey. 2007. Substitutions in the N-terminal alpha helical spine of Neisseria gonorrhoeae pilin affect type IV pilus assembly, dynamics and associated functions. Mol. Microbiol. 63:69-85. - PubMed
-
- Alm, R. A., and J. S. Mattick. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol. Microbiol. 16:485-496. - PubMed
-
- Alm, R. A., J. P. Hallinan, A. A. Watson, and J. S. Mattick. 1996. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol. Microbiol. 22:161-173. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

