Two essential arginine residues in the T components of energy-coupling factor transporters
- PMID: 19717603
- PMCID: PMC2795302
- DOI: 10.1128/JB.00965-09
Two essential arginine residues in the T components of energy-coupling factor transporters
Abstract
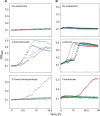
Energy-coupling factor (ECF) transporters, a recently discovered class of importers of micronutrients, are composed of a substrate-specific transmembrane component (S component) and a conserved energy-coupling module consisting of a transmembrane protein (T component) and pairs of ABC ATPases (A proteins). Based on utilization of a dedicated (subclass I) or shared (subclass II) energy-coupling module, ECF systems fall into two subclasses. The T components are the least-characterized proteins of ECF importers, and their function is essentially unknown. Using RcBioN and LmEcfT, the T units of the subclass I biotin transporter (RcBioMNY) of a gram-negative bacterium and of the subclass II folate, pantothenate, and riboflavin transporters of a lactic acid bacterium, respectively, we analyzed the role of two strongly conserved short motifs, each containing an arginine residue. Individual replacement of the two Arg residues in RcBioN reduced ATPase activity, an indicator of the transporter function, by two-thirds without affecting the modular assembly of the RcBioMNY complex. A double Arg-to-Glu replacement destroyed the complex and abolished ATPase activity. The corresponding single mutation in motif II of LmEcfT, as well as a double mutation, led to loss of the T unit from the subclass II ECF transporters and inactivated these systems. A single Arg-to-Glu replacement in motif I, however, abolished vitamin uptake activity without affecting assembly of the modules. Our results indicate that the conserved motif I in T components is essential for intramolecular signaling and, in cooperation with motif II, for subunit assembly of modular ECF transporters.
Figures
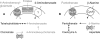


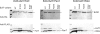

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

