Automated docking screens: a feasibility study
- PMID: 19719084
- PMCID: PMC2745826
- DOI: 10.1021/jm9006966
Automated docking screens: a feasibility study
Abstract
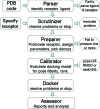
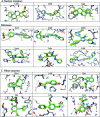
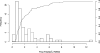
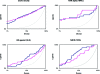
Molecular docking is the most practical approach to leverage protein structure for ligand discovery, but the technique retains important liabilities that make it challenging to deploy on a large scale. We have therefore created an expert system, DOCK Blaster, to investigate the feasibility of full automation. The method requires a PDB code, sometimes with a ligand structure, and from that alone can launch a full screen of large libraries. A critical feature is self-assessment, which estimates the anticipated reliability of the automated screening results using pose fidelity and enrichment. Against common benchmarks, DOCK Blaster recapitulates the crystal ligand pose within 2 A rmsd 50-60% of the time; inferior to an expert, but respectrable. Half the time the ligand also ranked among the top 5% of 100 physically matched decoys chosen on the fly. Further tests were undertaken culminating in a study of 7755 eligible PDB structures. In 1398 cases, the redocked ligand ranked in the top 5% of 100 property-matched decoys while also posing within 2 A rmsd, suggesting that unsupervised prospective docking is viable. DOCK Blaster is available at http://blaster.docking.org .
Figures


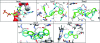

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

