RNA-based computation in live cells
- PMID: 19720518
- PMCID: PMC2764246
- DOI: 10.1016/j.copbio.2009.08.002
RNA-based computation in live cells
Abstract
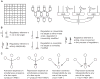
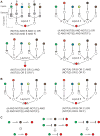
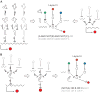
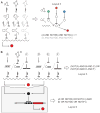
Man-made molecular 'computers' that operate inside live cells will enable unprecedented level of control over cellular physiology. A promising approach to building these computers uses RNA molecules and RNA-based regulation. RNA naturally lends itself to create 'digital' molecular networks that embody standardized (normal) forms of logic functions. The network's inputs, that may or may not be inverted by single-input NOT logic gates, feed into multi-input AND gates whose outputs are in turn integrated in a multi-input OR gate. Below I review recent steps that have been taken toward implementing these networks with allosteric riboswitches and ribozymes in bacteria and yeast, and RNAi in mammalian cells. I also propose how to co-opt recently discovered additional RNA regulation mechanisms into future construction efforts.
Figures
References
-
- Shapiro E, Benenson Y. Bringing DNA computers to life. Scientific American. 2006;295:44–51. - PubMed
-
- Stojanovic MN, Stefanovic D. A deoxyribozyme-based molecular automaton. Nature Biotechnology. 2003;21:1069–1074. - PubMed
-
- Benenson Y, Gil B, Ben-Dor U, Adar R, Shapiro E. An autonomous molecular computer for logical control of gene expression. Nature. 2004;429:423–429. - PubMed
-
- Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585–1588. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

