Severe insulin resistance and intrauterine growth deficiency associated with haploinsufficiency for INSR and CHN2: new insights into synergistic pathways involved in growth and metabolism
- PMID: 19720790
- PMCID: PMC2780873
- DOI: 10.2337/db09-0787
Severe insulin resistance and intrauterine growth deficiency associated with haploinsufficiency for INSR and CHN2: new insights into synergistic pathways involved in growth and metabolism
Abstract
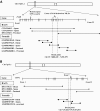
Objective: Digenic causes of human disease are rarely reported. Insulin via its receptor, which is encoded by INSR, plays a key role in both metabolic and growth signaling pathways. Heterozygous INSR mutations are the most common cause of monogenic insulin resistance. However, growth retardation is only reported with homozygous or compound heterozygous mutations. We describe a novel translocation [t(7,19)(p15.2;p13.2)] cosegregating with insulin resistance and pre- and postnatal growth deficiency. Chromosome translocations present a unique opportunity to identify modifying loci; therefore, our objective was to determine the mutational mechanism resulting in this complex phenotype.
Research design and methods: Breakpoint mapping was performed by fluorescence in situ hybridization (FISH) on patient chromosomes. Sequencing and gene expression studies of disrupted and adjacent genes were performed on patient-derived tissues. RESULTS Affected individuals had increased insulin, C-peptide, insulin-to-C-peptide ratio, and adiponectin levels consistent with an insulin receptoropathy. FISH mapping established that the translocation breakpoints disrupt INSR on chromosome 19p15.2 and CHN2 on chromosome 7p13.2. Sequencing demonstrated INSR haploinsufficiency accounting for elevated insulin levels and dysglycemia. CHN2 encoding beta-2 chimerin was shown to be expressed in insulin-sensitive tissues, and its disruption was shown to result in decreased gene expression in patient-derived adipose tissue.
Conclusions: We present a likely digenic cause of insulin resistance and growth deficiency resulting from the combined heterozygous disruption of INSR and CHN2, implicating CHN2 for the first time as a key element of proximal insulin signaling in vivo.
Figures



References
-
- Savage DB, Agostini M, Barroso I, Gurnell M, Luan J, Meirhaeghe A, Harding AH, Ihrke G, Rajanayagam O, Soos MA, George S, Berger D, Thomas EL, Bell JD, Meeran K, Ross RJ, Vidal-Puig A, Wareham NJ, O'Rahilly S, Chatterjee VK, Schafer AJ: Digenic inheritance of severe insulin resistance in a human pedigree. Nat Genet 2002; 31: 379– 384 - PubMed
-
- Taniguchi CM, Emanuelli B, Kahn CR: Critical nodes in signaling pathways: insights into insulin action. Nat Rev Mol Cell Biol 2006; 7: 85– 96 - PubMed
-
- Kahn CR, Flier JS, Bar RS, Archer JA, Gorden P, Martin MM, Roth J: The syndromes of insulin resistance and acanthosis nigricans: insulin-receptor disorders in man. N Engl J Med 1976; 294: 739– 745 - PubMed
-
- Krook A, Brueton L, O'Rahilly S: Homozygous nonsense mutation in the insulin receptor gene in infant with leprechaunism. Lancet 1993; 342: 277– 278 - PubMed
-
- Longo N, Wang Y, Smith SA, Langley SD, DiMeglio LA, Giannella-Neto D: Genotype-phenotype correlation in inherited severe insulin resistance. Hum Mol Genet 2002; 11: 1465– 1475 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

