Diversity, epidemiology, and genetics of class D beta-lactamases
- PMID: 19721065
- PMCID: PMC2798486
- DOI: 10.1128/AAC.01512-08
Diversity, epidemiology, and genetics of class D beta-lactamases
Abstract
Class D beta-lactamase-mediated resistance to beta-lactams has been increasingly reported during the last decade. Those enzymes also known as oxacillinases or OXAs are widely distributed among Gram negatives. Genes encoding class D beta-lactamases are known to be intrinsic in many Gram-negative rods, including Acinetobacter baumannii and Pseudomonas aeruginosa, but play a minor role in natural resistance phenotypes. The OXAs (ca. 150 variants reported so far) are characterized by an important genetic diversity and a great heterogeneity in terms of beta-lactam hydrolysis spectrum. The acquired OXAs possess either a narrow spectrum or an expanded spectrum of hydrolysis, including carbapenems in several instances. Acquired class D beta-lactamase genes are mostly associated to class 1 integron or to insertion sequences.
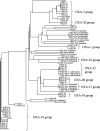
Figures

References
-
- Ambler, R. P. 1980. The structure of β-lactamases. Phil. Trans. R. Soc. London B Biol. Sci. 289:321-331. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

