Making the right connections: biological networks in the light of evolution
- PMID: 19722181
- PMCID: PMC2962804
- DOI: 10.1002/bies.200900043
Making the right connections: biological networks in the light of evolution
Abstract
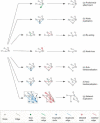
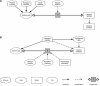
Our understanding of how evolution acts on biological networks remains patchy, as is our knowledge of how that action is best identified, modelled and understood. Starting with network structure and the evolution of protein-protein interaction networks, we briefly survey the ways in which network evolution is being addressed in the fields of systems biology, development and ecology. The approaches highlighted demonstrate a movement away from a focus on network topology towards a more integrated view, placing biological properties centre-stage. We argue that there remains great potential in a closer synergy between evolutionary biology and biological network analysis, although that may require the development of novel approaches and even different analogies for biological networks themselves.
Figures

Similar articles
-
Phylosystemics: Merging Phylogenomics, Systems Biology, and Ecology to Study Evolution.Trends Microbiol. 2020 Mar;28(3):176-190. doi: 10.1016/j.tim.2019.10.011. Epub 2019 Nov 28. Trends Microbiol. 2020. PMID: 31785987 Review.
-
Life's attractors : understanding developmental systems through reverse engineering and in silico evolution.Adv Exp Med Biol. 2012;751:93-119. doi: 10.1007/978-1-4614-3567-9_5. Adv Exp Med Biol. 2012. PMID: 22821455
-
Evolutionary principles underlying structure and response dynamics of cellular networks.Adv Exp Med Biol. 2012;751:225-47. doi: 10.1007/978-1-4614-3567-9_11. Adv Exp Med Biol. 2012. PMID: 22821461 Review.
-
Evolution of resource and energy management in biologically realistic gene regulatory network models.Adv Exp Med Biol. 2012;751:301-28. doi: 10.1007/978-1-4614-3567-9_14. Adv Exp Med Biol. 2012. PMID: 22821464
-
Organization principles in genetic interaction networks.Adv Exp Med Biol. 2012;751:53-78. doi: 10.1007/978-1-4614-3567-9_3. Adv Exp Med Biol. 2012. PMID: 22821453 Review.
Cited by
-
Unboxing cluster heatmaps.BMC Bioinformatics. 2017 Feb 15;18(Suppl 2):63. doi: 10.1186/s12859-016-1442-6. BMC Bioinformatics. 2017. PMID: 28251868 Free PMC article.
-
Systems Modeling at Multiple Levels of Regulation: Linking Systems and Genetic Networks to Spatially Explicit Plant Populations.Plants (Basel). 2013 Jan 25;2(1):16-49. doi: 10.3390/plants2010016. Plants (Basel). 2013. PMID: 27137364 Free PMC article. Review.
-
Linkage disequilibrium network analysis (LDna) gives a global view of chromosomal inversions, local adaptation and geographic structure.Mol Ecol Resour. 2015 Sep;15(5):1031-45. doi: 10.1111/1755-0998.12369. Epub 2015 Jan 21. Mol Ecol Resour. 2015. PMID: 25573196 Free PMC article.
-
EvoluCode: Evolutionary Barcodes as a Unifying Framework for Multilevel Evolutionary Data.Evol Bioinform Online. 2012;8:61-77. doi: 10.4137/EBO.S8814. Epub 2011 Dec 21. Evol Bioinform Online. 2012. PMID: 22267905 Free PMC article.
-
Evolution of protein-protein interaction networks in yeast.PLoS One. 2017 Mar 1;12(3):e0171920. doi: 10.1371/journal.pone.0171920. eCollection 2017. PLoS One. 2017. PMID: 28248977 Free PMC article.
References
-
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. - PubMed
-
- Dobzhansky T. Biology, molecular and organismic. Am Zool. 1964;4:443–452. - PubMed
-
- Schneeberger A, Mercer CH, Gregson SA, Ferguson NM, Nyamukapa CA, et al. Scale-free networks and sexually transmitted diseases: a description of observed patterns of sexual contacts in Britain and Zimbabwe. Sex Transm Dis. 2004;31:380–387. - PubMed
-
- Proulx SR, Promislow DEL, Phillips PC. Network thinking in ecology and evolution. Trends Ecol Evol. 2005;20:345–353. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

