Talking to chromatin: post-translational modulation of polycomb group function
- PMID: 19723311
- PMCID: PMC2745409
- DOI: 10.1186/1756-8935-2-10
Talking to chromatin: post-translational modulation of polycomb group function
Abstract
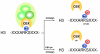
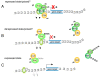
Polycomb Group proteins are important epigenetic regulators of gene expression. Epigenetic control by polycomb Group proteins involves intrinsic as well as associated enzymatic activities. Polycomb target genes change with cellular context, lineage commitment and differentiation status, revealing dynamic regulation of polycomb function. It is currently unclear how this dynamic modulation is controlled and how signaling affects polycomb-mediated epigenetic processes at the molecular level. Experimental evidence on regulation of polycomb function by post-translational mechanisms is steadily emerging: Polycomb Group proteins are targeted for ubiquitylation, sumoylation and phosphorylation. In addition, specific Polycomb Group proteins modify other (chromatin) associated proteins via similar post-translational modifications. Such modifications affect protein function by affecting protein stability, protein-protein interactions and enzymatic activities. Here, we review current insights in covalent modification of Polycomb Group proteins in the context of protein function and present a tentative view of integrated signaling to chromatin in the context of phosphorylation. Clearly, the available literature reveals just the tip of the iceberg, and exact molecular mechanisms in, and the biological relevance of post-translational regulation of polycomb function await further elucidation. Our understanding of causes and consequences of post-translational modification of polycomb proteins will gain significantly from in vivo validation experiments. Impaired polycomb function has important repercussions for stem cell function, development and disease. Ultimately, increased understanding of signaling to chromatin and the mechanisms involved in epigenetic remodeling will contribute to the development of therapeutic interventions in cell fate decisions in development and disease.
Figures


Similar articles
-
The polycomb repressive complex 2 is a potential target of SUMO modifications.PLoS One. 2008 Jul 16;3(7):e2704. doi: 10.1371/journal.pone.0002704. PLoS One. 2008. PMID: 18628979 Free PMC article.
-
Concise review: roles of polycomb group proteins in development and disease: a stem cell perspective.Stem Cells. 2007 Oct;25(10):2498-510. doi: 10.1634/stemcells.2006-0608. Epub 2007 Jun 28. Stem Cells. 2007. PMID: 17600113 Review.
-
Pancreas development and the Polycomb group protein complexes.Mech Dev. 2020 Dec;164:103647. doi: 10.1016/j.mod.2020.103647. Epub 2020 Sep 28. Mech Dev. 2020. PMID: 32991980 Review.
-
MMTR/Dmap1 Sets the Stage for Early Lineage Commitment of Embryonic Stem Cells by Crosstalk with PcG Proteins.Cells. 2020 May 11;9(5):1190. doi: 10.3390/cells9051190. Cells. 2020. PMID: 32403252 Free PMC article.
-
Cell Fate and Developmental Regulation Dynamics by Polycomb Proteins and 3D Genome Architecture.Bioessays. 2019 Mar;41(3):e1800222. doi: 10.1002/bies.201800222. Epub 2019 Feb 22. Bioessays. 2019. PMID: 30793782 Review.
Cited by
-
The HDAC inhibitor SAHA regulates CBX2 stability via a SUMO-triggered ubiquitin-mediated pathway in leukemia.Oncogene. 2018 May;37(19):2559-2572. doi: 10.1038/s41388-018-0143-1. Epub 2018 Feb 22. Oncogene. 2018. PMID: 29467492 Free PMC article.
-
LymphoAtlas: a dynamic and integrated phosphoproteomic resource of TCR signaling in primary T cells reveals ITSN2 as a regulator of effector functions.Mol Syst Biol. 2020 Jul;16(7):e9524. doi: 10.15252/msb.20209524. Mol Syst Biol. 2020. PMID: 32618424 Free PMC article.
-
Mitogen-activated protein kinase signaling mediates phosphorylation of polycomb ortholog Cbx7.J Biol Chem. 2013 Dec 20;288(51):36398-408. doi: 10.1074/jbc.M113.486266. Epub 2013 Nov 5. J Biol Chem. 2013. PMID: 24194518 Free PMC article.
-
Quantitative in vivo analysis of chromatin binding of Polycomb and Trithorax group proteins reveals retention of ASH1 on mitotic chromatin.Nucleic Acids Res. 2013 May 1;41(10):5235-50. doi: 10.1093/nar/gkt217. Epub 2013 Apr 10. Nucleic Acids Res. 2013. PMID: 23580551 Free PMC article.
-
Stuxnet Facilitates the Degradation of Polycomb Protein during Development.Dev Cell. 2016 Jun 20;37(6):507-19. doi: 10.1016/j.devcel.2016.05.013. Dev Cell. 2016. PMID: 27326929 Free PMC article.
References
LinkOut - more resources
Full Text Sources

