The B6 database: a tool for the description and classification of vitamin B6-dependent enzymatic activities and of the corresponding protein families
- PMID: 19723314
- PMCID: PMC2748086
- DOI: 10.1186/1471-2105-10-273
The B6 database: a tool for the description and classification of vitamin B6-dependent enzymatic activities and of the corresponding protein families
Abstract
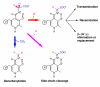
Background: Enzymes that depend on vitamin B6 (and in particular on its metabolically active form, pyridoxal 5'-phosphate, PLP) are of great relevance to biology and medicine, as they catalyze a wide variety of biochemical reactions mainly involving amino acid substrates. Although PLP-dependent enzymes belong to a small number of independent evolutionary lineages, they encompass more than 160 distinct catalytic functions, thus representing a striking example of divergent evolution. The importance and remarkable versatility of these enzymes, as well as the difficulties in their functional classification, create a need for an integrated source of information about them.
Description: The B6 database http://bioinformatics.unipr.it/B6db contains documented B6-dependent activities and the relevant protein families, defined as monophyletic groups of sequences possessing the same enzymatic function. One or more families were associated to each of 121 PLP-dependent activities with known sequences. Hidden Markov models (HMMs) were built from family alignments and incorporated in the database. These HMMs can be used for the functional classification of PLP-dependent enzymes in genomic sets of predicted protein sequences. An example of such analyses (a census of human genes coding for PLP-dependent enzymes) is provided here, whereas many more are accessible through the database itself.
Conclusion: The B6 database is a curated repository of biochemical and molecular information about an important group of enzymes. This information is logically organized and available for computational analyses, providing a key resource for the identification, classification and comparative analysis of B6-dependent enzymes.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

